Gov NonGov
0 1 Generalized Measurement Models: Modeling Multidimensional Latent Variables
Today’s Lecture Objectives
- Show how to estimate multidimensional latent variable models with polytomous data
Example Data: Conspiracy Theories
Today’s example is from a bootstrap resample of 177 undergraduate students at a large state university in the Midwest. The survey was a measure of 10 questions about their beliefs in various conspiracy theories that were being passed around the internet in the early 2010s. Additionally, gender was included in the survey. All items responses were on a 5- point Likert scale with:
- Strongly Disagree
- Disagree
- Neither Agree or Disagree
- Agree
- Strongly Agree
Please note, the purpose of this survey was to study individual beliefs regarding conspiracies. The questions can provoke some strong emotions given the world we live in currently. All questions were approved by university IRB prior to their use.
Our purpose in using this instrument is to provide a context that we all may find relevant as many of these conspiracy theories are still prevalent today.
Conspiracy Theory Questions 1-5
Questions:
- The U.S. invasion of Iraq was not part of a campaign to fight terrorism, but was driven by oil companies and Jews in the U.S. and Israel.
- Certain U.S. government officials planned the attacks of September 11, 2001 because they wanted the United States to go to war in the Middle East.
- President Barack Obama was not really born in the United States and does not have an authentic Hawaiian birth certificate.
- The current financial crisis was secretly orchestrated by a small group of Wall Street bankers to extend the power of the Federal Reserve and further their control of the world’s economy.
- Vapor trails left by aircraft are actually chemical agents deliberately sprayed in a clandestine program directed by government officials.
Conspiracy Theory Questions 6-10
Questions:
- Billionaire George Soros is behind a hidden plot to destabilize the American government, take control of the media, and put the world under his control.
- The U.S. government is mandating the switch to compact fluorescent light bulbs because such lights make people more obedient and easier to control.
- Government officials are covertly Building a 12-lane "NAFTA superhighway" that runs from Mexico to Canada through America’s heartland.
- Government officials purposely developed and spread drugs like crack-cocaine and diseases like AIDS in order to destroy the African American community.
- God sent Hurricane Katrina to punish America for its sins.
Latent Variables
The Latent Variables
Latent variables are built by specification:
- What is their distribution? (nearly all are normally distributed)
- How do you specify their scale: mean and standard deviation? (step two in our model analysis steps)
You create latent variables by specifying which items measure which latent variables in an analysis model
- Called different names by different fields:
- Alignment (educational measurement)
- Factor pattern matrix (factor analysis)
- Q-matrix (Question matrix; diagnostic models and multidimensional IRT)
From Q-matrix to Model
The alignment provides a specification of which latent variables are measured by which items
- Sometimes we say items “load onto” factors
The mathematical definition of either of these terms is simply whether or not a latent variable appears as a predictor for an item
- For instance, item one appears to measure nongovernment conspiracies, meaning its alignment (row vector of the Q-matrix) would be:
The model for the first item is then built with only the factors measured by the item as being present:
\[ f(E\left(Y_{p1} \mid \boldsymbol{\theta}_p\right) ) = \mu_1 + \lambda_{11} \theta_{p1} \]
From Q-matrix to Model
The model for the first item is then built with only the factors measured by the item as being present:
\[ f(E\left(Y_{p1} \mid \boldsymbol{\theta}_p\right) ) = \mu_1 + \lambda_{11} \theta_{p1} \]
Where:
- \(\mu_1\) is the item intercept
- \(\lambda_{11}\) is the factor loading for item 1 (the first subscript) for factor 1 (the second subscript)
- \(\theta_{p1}\) is the value of the latent variable for person \(p\) and factor 1
The second factor is not included in the model for the item.
More on the Q-matrix
We could show the model with the Q-matrix entries:
\[ f(E\left(Y_{p1} \mid \boldsymbol{\theta}_p\right) ) = \mu_1 + q_{11}\left( \lambda_{11} \theta_{p1} \right) + q_{12}\left( \lambda_{12} \theta_{p2} \right) = \mu_1 + \boldsymbol{\theta}_p \text{diag}\left(\boldsymbol{q}_i \right) \boldsymbol{\lambda}_{1} \]
Where:
\(\boldsymbol{\lambda}_{1} = \left[ \begin{array}{c} \lambda_{11} \\ \lambda_{12} \end{array} \right]\) contains all possible factor loadings for item 1 (size \(2 \times 1\))
\(\boldsymbol{\theta}_p = \left[ \begin{array}{cc} \theta_{p1} & \theta_{p2} \\ \end{array} \right]\) contains the factor scores for person \(p\) (size \(1 \times 2\))
\(\text{diag}\left(\boldsymbol{q}_i \right) = \boldsymbol{q}_i \left[ \begin{array}{cc} 1 & 0 \\ 0 & 1 \end{array} \right] = \left[ \begin{array}{cc} 0 & 1 \\ \end{array} \right] \left[ \begin{array}{cc} 1 & 0 \\ 0 & 1 \end{array} \right] = \left[ \begin{array}{cc} 0 & 0 \\ 0 & 1 \end{array} \right]\) is a diagonal matrix of ones times the vector of Q-matrix entries for item 1
Even More on the Q-matrix
The matrix product then gives:
\[\boldsymbol{\theta}_p \text{diag}\left(\boldsymbol{q}_i \right) \boldsymbol{\lambda}_{1} = \left[ \begin{array}{cc} \theta_{p1} & \theta_{p2} \\ \end{array} \right]\left[ \begin{array}{cc} 0 & 0 \\ 0 & 1 \end{array} \right] \left[ \begin{array}{c} \lambda_{11} \\ \lambda_{12} \end{array} \right] = \left[ \begin{array}{cc} \theta_{p1} & \theta_{p2} \\ \end{array} \right]\left[ \begin{array}{c} 0 \\ \lambda_{12} \end{array} \right] = \lambda_{12}\theta_{p2}\]
The Q-matrix functions like a partial version of the model (predictor) matrix that we saw in linear models
Today’s Q-matrix
Gov NonGov
item1 0 1
item2 1 0
item3 0 1
item4 0 1
item5 1 0
item6 0 1
item7 1 0
item8 1 0
item9 1 0
item10 0 1Measurement Model Implied by Today’s Q-matrix
Given the Q-matrix each item has its own model using the alignment specifications
\[ \begin{array}{c} f(E\left(Y_{p1} \mid \boldsymbol{\theta}_p\right) ) = \mu_1 + \lambda_{12} \theta_{p2} \\ f(E\left(Y_{p2} \mid \boldsymbol{\theta}_p\right) ) = \mu_2 + \lambda_{21} \theta_{p1} \\ f(E\left(Y_{p3} \mid \boldsymbol{\theta}_p\right) ) = \mu_3 + \lambda_{32} \theta_{p2} \\ f(E\left(Y_{p4} \mid \boldsymbol{\theta}_p\right) ) = \mu_4 + \lambda_{42} \theta_{p2} \\ f(E\left(Y_{p5} \mid \boldsymbol{\theta}_p\right) ) = \mu_5 + \lambda_{51} \theta_{p1} \\ f(E\left(Y_{p6} \mid \boldsymbol{\theta}_p\right) ) = \mu_6 + \lambda_{62} \theta_{p2} \\ f(E\left(Y_{p7} \mid \boldsymbol{\theta}_p\right) ) = \mu_7 + \lambda_{71} \theta_{p1} \\ f(E\left(Y_{p8} \mid \boldsymbol{\theta}_p\right) ) = \mu_8 + \lambda_{81} \theta_{p1} \\ f(E\left(Y_{p9} \mid \boldsymbol{\theta}_p\right) ) = \mu_9 + \lambda_{91} \theta_{p1} \\ f(E\left(Y_{p,10} \mid \boldsymbol{\theta}_p\right) ) = \mu_{10} + \lambda_{10,2} \theta_{p2} \\ \end{array} \]
Our Example: Multivariate Normal Distribution
For our example, we will assume the set of traits follows a multivariate normal distribution
\[ f\left(\boldsymbol{\theta}_p \right) = \left(2 \pi \right)^{-\frac{D}{2}} \det\left(\boldsymbol{\Sigma}_\theta \right)^{-\frac{1}{2}}\exp\left[-\frac{1}{2}\left(\boldsymbol{\theta}_p - \boldsymbol{\mu}_\theta \right)^T\boldsymbol{\Sigma}_\theta^{-1}\left(\boldsymbol{\theta}_p - \boldsymbol{\mu}_\theta \right) \right] \] Where:
- \(\pi \approx 3.14\)
- \(D\) is the number of latent variables (dimensions)
- \(\boldsymbol{\Sigma}_\theta\) is the covariance matrix of the latent variables
- \(\boldsymbol{\Sigma}_\theta^{-1}\) is the inverse of the covariance matrix
- \(\boldsymbol{\mu}_\theta\) is the mean vector of the latent variables
- \(\det\left( \cdot\right)\) is the matrix determinant function
- \(\left(\cdot \right)^T\) is the matrix transpose operator
Alternatively, we would specify \(\boldsymbol{\theta}_p \sim N_D\left( \boldsymbol{\mu}_\theta, \boldsymbol{\Sigma}_\theta \right)\); but, we cannot always estimate \(\boldsymbol{\mu}_\theta\) and \(\boldsymbol{\Sigma}_\theta\)
Identification of Latent Traits, Part 1
Psychometric models require two types of identification to be valid:
- Empirical Identification
- The minimum number of items that must measure each latent variable
- From CFA: three observed variables for each latent variable (or two if the latent variable is correlated with another latent variable)
Bayesian priors can help to make models with fewer items than these criteria suggest estimable
- The parameter estimates (item parameters and latent variable estimates) often have MCMC convergence issues and should not be trusted
- Use the CFA standard in your work
Identification of Latent Traits, Part 2
Psychometric models require two types of identification to be valid:
- Scale Identification (i.e., what the mean/variance is for each latent variable)
- The additional set of constraints needed to set the mean and standard deviation (variance) of the latent variables
- Two main methods to set the scale:
- Marker item parameters
- For variances: Set the loading/slope to one for one observed variable per latent variable
- Can estimate the latent variable’s variance (the diagonal of \(\boldsymbol{\Sigma}_\theta\))
- For means: Set the item intercept to one for one observed variable perlatent variable
- Can estimate the latent variable’s mean (in \(\boldsymbol{\mu}_\theta\))
- For variances: Set the loading/slope to one for one observed variable per latent variable
- Standardized factors
- Set the variance for all latent variables to one
- Set the mean for all latent variables to zero
- Estimate all unique off-diagonal correlations (covariances) in \(\boldsymbol{\Sigma}_\theta\)
- Marker item parameters
More on Scale Identification
Bayesian priors can let you believe you can estimate more parameters than the non-Bayesian standards suggest
- For instance, all item parameters and the latent variable means/variances
Like empirical identification, these estimates are often unstable and are not recommended
Most common:
- Standardized latent variables
- Used for scale development and/or when scores are of interest directly
- Marker item for latent variables and zero means
- Used for cases where latent variables may become outcomes (and that variance needs explained)
Today, we will use standardized factors
- Covariance matrix diagonal (factor variances) all ones (factor correlation matrix)
Observed Item Model
For each item, we will use a graded- (ordinal logit) response modelwhere:
\[P\left(Y_{ic } = c \mid \boldsymbol{\theta}_p \right) = \left\{ \begin{array}{lr} 1-P^*\left(Y_{i1} \gt 1 \mid \boldsymbol{\theta}_p \right) & \text{if } c=1 \\ P^*\left(Y_{i{c-1}} \gt c-1 \mid \boldsymbol{\theta}_p \right) - P^*\left(Y_{i{c}} \gt c \mid \boldsymbol{\theta}_p \right) & \text{if } 1<c<C_i \\ P^*\left(Y_{i{C_i -1} } \gt C_i-1 \mid \boldsymbol{\theta}_p \right) - 0 & \text{if } c=C_i \\ \end{array} \right.\]
Where:
\[ P^*\left(Y_{i{c}} > c \mid \theta \right) = \frac{\exp(\mu_{ic}+\boldsymbol{\theta}_p \text{diag}\left(\boldsymbol{q}_i \right) \boldsymbol{\lambda}_{i})} {1+\exp(\mu_{ic}+\boldsymbol{\theta}_p \text{diag}\left(\boldsymbol{q}_i \right) \boldsymbol{\lambda}_{i})}\]
With:
- \(C_i-1\) Ordered intercepts: \(\mu_1 > \mu_2 > \ldots > \mu_{C_i-1}\)
Building the Multidimensional Model in Stan
Observed Item Model in Stan
As Stan uses thresholds instead of intercepts, our model then becomes:
\[P\left(Y_{ic } = c \mid \boldsymbol{\theta}_p \right) = \left\{ \begin{array}{lr} 1-P\left(Y_{i1} \gt 1 \mid \boldsymbol{\theta}_p \right) & \text{if } c=1 \\ P\left(Y_{i{c-1}} \gt c-1 \mid \boldsymbol{\theta}_p \right) - P\left(Y_{i{c}} \gt c \mid \boldsymbol{\theta}_p \right) & \text{if } 1<c<C_i \\ P\left(Y_{i{C_i -1} } \gt C_i-1 \mid \boldsymbol{\theta}_p \right) & \text{if } c=C_i \\ \end{array} \right.\]
Where:
\[ P\left(Y_{i{c}} > c \mid \theta \right) = \frac{\exp(-\tau_{ic}+\boldsymbol{\theta}_p \text{diag}\left(\boldsymbol{q}_i \right) \boldsymbol{\lambda}_{i})} {1+\exp(-\tau_{ic}+\boldsymbol{\theta}_p \text{diag}\left(\boldsymbol{q}_i \right) \boldsymbol{\lambda}_{i})}\]
With:
- \(C_i-1\) Ordered thresholds: \(\tau_1 < \tau_2 < \ldots < \tau_{C_i-1}\)
We can convert thresholds to intercepts by multiplying by negative one: \(\mu_c = -\tau_c\)
Stan Model Block
model {
lambda ~ multi_normal(meanLambda, covLambda);
thetaCorrL ~ lkj_corr_cholesky(1.0);
theta ~ multi_normal_cholesky(meanTheta, thetaCorrL);
for (item in 1:nItems){
thr[item] ~ multi_normal(meanThr[item], covThr[item]);
Y[item] ~ ordered_logistic(thetaMatrix*lambdaMatrix[item,1:nFactors]', thr[item]);
}
}thetaMatrixis matrix of latent variables for each person \(\boldsymbol{\Theta}\) (size \(N \times D\); \(N\) is number of people, \(D\) is number of dimensions)lambdaMatrixis, for each item, a matrix of factor loading/discrimination parameters along with zeros by Q-matrix (size \(I \times D\); \(I\) is number of items)- The transpose of one row of the lambda matrix is used, resulting in a scalar for each person
lambdais a vector of estimated factor loadings (we use a different block to put these intolambdaMatrix)- Each factor loading has a (univariate) normal distribution for a prior
More Model Block Notes:
model {
lambda ~ multi_normal(meanLambda, covLambda);
thetaCorrL ~ lkj_corr_cholesky(1.0);
theta ~ multi_normal_cholesky(meanTheta, thetaCorrL);
for (item in 1:nItems){
thr[item] ~ multi_normal(meanThr[item], covThr[item]);
Y[item] ~ ordered_logistic(thetaMatrix*lambdaMatrix[item,1:nFactors]', thr[item]);
}
}thetais an array of latent variables (we use a different block to put these intothetaMatrix)thetafollows a multivariate normal distribution as a prior where:- We set all means of
thetato zero (the mean for each factor is set to zero) - We set all variances of
thetato one (the variance for each factor is set to one) - We estimate a correlation matrix of factors \(\textbf{R}_\theta\) using a so-called LKJ prior distribution
- LKJ priors are found in Stan and make modeling correlations very easy
- We set all means of
- Of note: we are using the Cholesky forms of the functions
lkj_corr_choleskyandmulti_normal_cholesky
LKJ Priors for Correlation Matrices
From Stan’s Functions Reference, for a correlation matrix \(\textbf{R}_\theta\)
Correlation Matrix Properties:
- Positive definite (determinant greater than zero; \(\det\left(R_\theta \right) >0\)
- Symmetric
- Diagonal values are all one
LKJ Prior, with hyperparameter \(\eta\), is proportional to the determinant of the correlation matrix
\[\text{LKJ}\left(\textbf{R}_\theta \mid \eta \right) \propto \det\left(\textbf{R}_\theta \right)^{(\eta-1)} \]
- Density is uniform over correlation matrices with \(\eta=1\)
- With \(\eta>1\), identity matrix is modal (moves correlations toward zero)
- With \(0<\eta<1\), density has a trough at the identity (moves correlations away from zero)
For this example, we set \(\eta=1\), noting a uniform prior over all correlation matrices
Cholesky Decomposition
The functions we are using do not use the correlation matrix directly
The problem:
- The issue: We need the inverse and determinant to calculate the density of a multivariate normal distribution
- Inverses and determinants are numerically unstable (computationally) in general form
The solution:
- Convert \(R_\theta\) to a triangular form using the Cholesky decomposition
\[\textbf{R}_\theta = \textbf{L}_\theta \textbf{L}_\theta^T\]
Cholesky Decomposition Math
- \(\det\left(\textbf{R}_\theta \right) = \det\left(\textbf{L}_\theta \textbf{L}_\theta^T\right) = \det\left(\textbf{L}_\theta\right) \det\left(\textbf{L}_\theta^T\right) = \det\left(\textbf{L}_\theta\right)^2 = \left(\prod_{d=1}^D l_{d,d}\right)^2\) (note: product becomes sum for log-likelihood)
- For the inverse, we work with the term in the exponent of the MVN pdf:
\[-\frac{1}{2}\left(\boldsymbol{\theta}_p - \boldsymbol{\mu}_\theta \right)^T\textbf{R}_\theta^{-1}\left(\boldsymbol{\theta}_p - \boldsymbol{\mu}_\theta \right) = -\frac{1}{2}\left(\boldsymbol{\theta}_p - \boldsymbol{\mu}_\theta \right)^T\left(\textbf{L}_\theta \textbf{L}_\theta^T\right)^{-1}\left(\boldsymbol{\theta}_p - \boldsymbol{\mu}_\theta \right) \]
\[ -\frac{1}{2}\left(\boldsymbol{\theta}_p - \boldsymbol{\mu}_\theta \right)^T\textbf{L}_\theta^{-T} \textbf{L}_\theta^{-1}\left(\boldsymbol{\theta}_p - \boldsymbol{\mu}_\theta \right)\] Then, we solve by back substitution: \(\left(\boldsymbol{\theta}_p - \boldsymbol{\mu}_\theta \right)^T\textbf{L}_\theta^{-T}\)
- We can then multiply this result by itself to form the term in the exponent (times \(-\frac{1}{2}\))
- Back substitution involves far fewer steps, minimizing the amount of rounding error in the process of forming the inverse
- Most algorithms using MVN distributions use some variant of this process (perhaps with a different factorization method such as QR)
Stan Parameters Block
parameters {
array[nObs] vector[nFactors] theta; // the latent variables (one for each person)
array[nItems] ordered[maxCategory-1] thr; // the item thresholds (one for each item category minus one)
vector[nLoadings] lambda; // the factor loadings/item discriminations (one for each item)
cholesky_factor_corr[nFactors] thetaCorrL;
}Notes:
thetais an array (for the MVN prior)thris the same as the unidimensional modellambdais the vector of all factor loadings to be estimated (needsnLoadings)thetaCorrLis of typecholesky_factor_corr, a built in type that identifies this as the lower diagonal of a Cholesky-factorized correlation matrix
We need a couple other blocks to link our parameters to the model
Stan Transformed Data Block
transformed data{
int<lower=0> nLoadings = 0; // number of loadings in model
for (factor in 1:nFactors){
nLoadings = nLoadings + sum(Qmatrix[1:nItems, factor]);
}
array[nLoadings, 2] int loadingLocation; // the row/column positions of each loading
int loadingNum=1;
for (item in 1:nItems){
for (factor in 1:nFactors){
if (Qmatrix[item, factor] == 1){
loadingLocation[loadingNum, 1] = item;
loadingLocation[loadingNum, 2] = factor;
loadingNum = loadingNum + 1;
}
}
}
}- The
transformed data {}block runs prior to the Markov chain- We use it to create variables that will stay constant throughout the chain
- This syntax works for any Q-matrix (but only has main effects in the model)
Stan Transformed Data Block
transformed data{
int<lower=0> nLoadings = 0; // number of loadings in model
for (factor in 1:nFactors){
nLoadings = nLoadings + sum(Qmatrix[1:nItems, factor]);
}
array[nLoadings, 2] int loadingLocation; // the row/column positions of each loading
int loadingNum=1;
for (item in 1:nItems){
for (factor in 1:nFactors){
if (Qmatrix[item, factor] == 1){
loadingLocation[loadingNum, 1] = item;
loadingLocation[loadingNum, 2] = factor;
loadingNum = loadingNum + 1;
}
}
}
}- Here, we count the number of loadings in the Q-matrix
nLoadings- We then process the Q-matrix to tell Stan the row and column position of each loading in the
loadingsMatrixused in themodel {}block - For instance, the loading for item one factor two has row position 1 and column position 2
- We then process the Q-matrix to tell Stan the row and column position of each loading in the
Stan Transformed Parameters Block
transformed parameters{
matrix[nItems, nFactors] lambdaMatrix = rep_matrix(0.0, nItems, nFactors);
matrix[nObs, nFactors] thetaMatrix;
// build matrix for lambdas to multiply theta matrix
for (loading in 1:nLoadings){
lambdaMatrix[loadingLocation[loading,1], loadingLocation[loading,2]] = lambda[loading];
}
for (factor in 1:nFactors){
thetaMatrix[,factor] = to_vector(theta[,factor]);
}
}Notes:
- The
transformed parameters {}block runs prior to each iteration of the Markov chain- This means it affects the estimation of each type of parameter
- We use it to create:
thetaMatrix(convertingthetafrom an array to a matrix)lambdaMatrix(puts the loadings and zeros from the Q-matrix into correct position)lambdaMatrixinitializes at zero (so we just have to add the loadings in the correct position)
Stan Data Block
data {
// data specifications =============================================================
int<lower=0> nObs; // number of observations
int<lower=0> nItems; // number of items
int<lower=0> maxCategory; // number of categories for each item
// input data =============================================================
array[nItems, nObs] int<lower=1, upper=5> Y; // item responses in an array
// loading specifications =============================================================
int<lower=1> nFactors; // number of loadings in the model
array[nItems, nFactors] int<lower=0, upper=1> Qmatrix;
// prior specifications =============================================================
array[nItems] vector[maxCategory-1] meanThr; // prior mean vector for intercept parameters
array[nItems] matrix[maxCategory-1, maxCategory-1] covThr; // prior covariance matrix for intercept parameters
vector[nItems] meanLambda; // prior mean vector for discrimination parameters
matrix[nItems, nItems] covLambda; // prior covariance matrix for discrimination parameters
vector[nFactors] meanTheta;
}- Changes from unidimensional model:
meanTheta: Factor means (hyperparameters) are added (but we will set these to zero)nFactors: Number of latent variables (needed for Q-matrix)Qmatrix: Q-matrix for model
Stan Generated Quantities Block
Notes:
- Converts thresholds to intercepts
mu - Creates
thetaCorrby multiplying Cholesky-factorized lower triangle with upper triangle- We will only use the parameters of
thetaCorrwhen looking at model output
- We will only use the parameters of
Stan Analyses
Estimation in Stan
We run stan the same way we have previously:
Notes:
- Smaller chain (model takes a lot longer to run)
- Still need to keep loadings positive initially
Analysis Results
[1] 1.066736# A tibble: 54 × 10
variable mean median sd mad q5 q95 rhat ess_bulk
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 lambda[1] 1.99 1.98 0.266 0.265 1.58 2.45 1.00 2970.
2 lambda[2] 2.83 2.81 0.383 0.383 2.24 3.49 1.00 2898.
3 lambda[3] 2.40 2.38 0.341 0.333 1.88 2.99 1.00 3532.
4 lambda[4] 2.92 2.89 0.397 0.386 2.32 3.61 1.00 2965.
5 lambda[5] 4.35 4.30 0.594 0.578 3.44 5.38 1.00 2501.
6 lambda[6] 4.22 4.18 0.565 0.551 3.37 5.19 1.00 2569.
7 lambda[7] 2.85 2.83 0.407 0.405 2.21 3.55 1.00 3043.
8 lambda[8] 4.11 4.08 0.557 0.549 3.25 5.09 1.00 2468.
9 lambda[9] 2.91 2.89 0.429 0.425 2.24 3.64 1.00 3174.
10 lambda[10] 2.44 2.41 0.424 0.424 1.79 3.17 1.00 4309.
11 mu[1,1] 1.88 1.88 0.288 0.283 1.41 2.37 1.00 1579.
12 mu[2,1] 0.815 0.809 0.300 0.299 0.329 1.31 1.00 946.
13 mu[3,1] 0.100 0.104 0.261 0.257 -0.337 0.530 1.00 974.
14 mu[4,1] 0.998 0.990 0.318 0.310 0.481 1.54 1.00 922.
15 mu[5,1] 1.32 1.32 0.425 0.426 0.651 2.04 1.00 766.
16 mu[6,1] 0.976 0.969 0.401 0.405 0.318 1.64 1.00 749.
17 mu[7,1] -0.102 -0.100 0.291 0.294 -0.577 0.371 1.00 851.
18 mu[8,1] 0.699 0.691 0.388 0.384 0.0691 1.35 1.00 766.
19 mu[9,1] -0.0597 -0.0602 0.295 0.290 -0.548 0.431 1.00 888.
20 mu[10,1] -1.36 -1.34 0.309 0.302 -1.90 -0.881 1.00 1297.
21 mu[1,2] -0.208 -0.206 0.230 0.231 -0.593 0.168 1.00 1104.
22 mu[2,2] -1.52 -1.51 0.319 0.316 -2.06 -1.01 1.00 1024.
23 mu[3,2] -1.11 -1.10 0.272 0.270 -1.57 -0.670 1.00 1090.
24 mu[4,2] -1.14 -1.13 0.314 0.310 -1.68 -0.643 1.00 977.
25 mu[5,2] -1.97 -1.95 0.448 0.451 -2.75 -1.28 1.00 966.
26 mu[6,2] -1.99 -1.98 0.426 0.427 -2.72 -1.32 1.01 905.
27 mu[7,2] -1.94 -1.93 0.336 0.328 -2.53 -1.41 1.00 1184.
28 mu[8,2] -1.82 -1.81 0.411 0.405 -2.51 -1.16 1.00 1028.
29 mu[9,2] -1.95 -1.94 0.345 0.344 -2.55 -1.42 1.00 1204.
30 mu[10,2] -2.60 -2.58 0.368 0.356 -3.24 -2.03 1.00 1879.
31 mu[1,3] -2.03 -2.03 0.275 0.274 -2.50 -1.59 1.00 1467.
32 mu[2,3] -3.38 -3.36 0.411 0.409 -4.08 -2.75 1.00 1787.
33 mu[3,3] -3.68 -3.66 0.433 0.428 -4.44 -3.01 1.00 2767.
34 mu[4,3] -3.85 -3.84 0.462 0.461 -4.64 -3.13 1.00 1881.
35 mu[5,3] -4.59 -4.55 0.609 0.589 -5.64 -3.66 1.00 1717.
36 mu[6,3] -5.63 -5.59 0.681 0.683 -6.79 -4.57 1.00 2624.
37 mu[7,3] -4.14 -4.11 0.498 0.494 -5.01 -3.39 1.00 2239.
38 mu[8,3] -6.42 -6.38 0.769 0.763 -7.76 -5.22 1.00 4332.
39 mu[9,3] -3.25 -3.24 0.415 0.408 -3.97 -2.60 1.00 1519.
40 mu[10,3] -3.77 -3.74 0.464 0.459 -4.56 -3.05 1.00 2408.
41 mu[1,4] -3.98 -3.96 0.464 0.456 -4.77 -3.25 1.00 4398.
42 mu[2,4] -4.92 -4.89 0.580 0.577 -5.92 -4.02 1.00 3244.
43 mu[3,4] -4.78 -4.76 0.555 0.551 -5.73 -3.92 1.00 4647.
44 mu[4,4] -4.76 -4.74 0.547 0.542 -5.71 -3.92 1.00 2442.
45 mu[5,4] -6.86 -6.82 0.841 0.836 -8.32 -5.57 1.00 3061.
46 mu[6,4] -7.95 -7.91 0.967 0.982 -9.62 -6.45 1.00 4485.
47 mu[7,4] -5.70 -5.67 0.690 0.685 -6.86 -4.62 1.00 4292.
48 mu[8,4] -8.47 -8.40 1.08 1.09 -10.4 -6.82 1.00 5871.
49 mu[9,4] -4.95 -4.92 0.584 0.569 -5.97 -4.05 1.00 2476.
50 mu[10,4] -4.02 -4.00 0.490 0.490 -4.87 -3.26 1.00 2909.
51 thetaCorr[1,1] 1 1 0 0 1 1 NA NA
52 thetaCorr[2,1] 0.990 0.992 0.00742 0.00672 0.976 0.999 1.07 72.0
53 thetaCorr[1,2] 0.990 0.992 0.00742 0.00672 0.976 0.999 1.07 72.0
54 thetaCorr[2,2] 1 1 0 0 1 1 NA NA
# ℹ 1 more variable: ess_tail <dbl>Note:
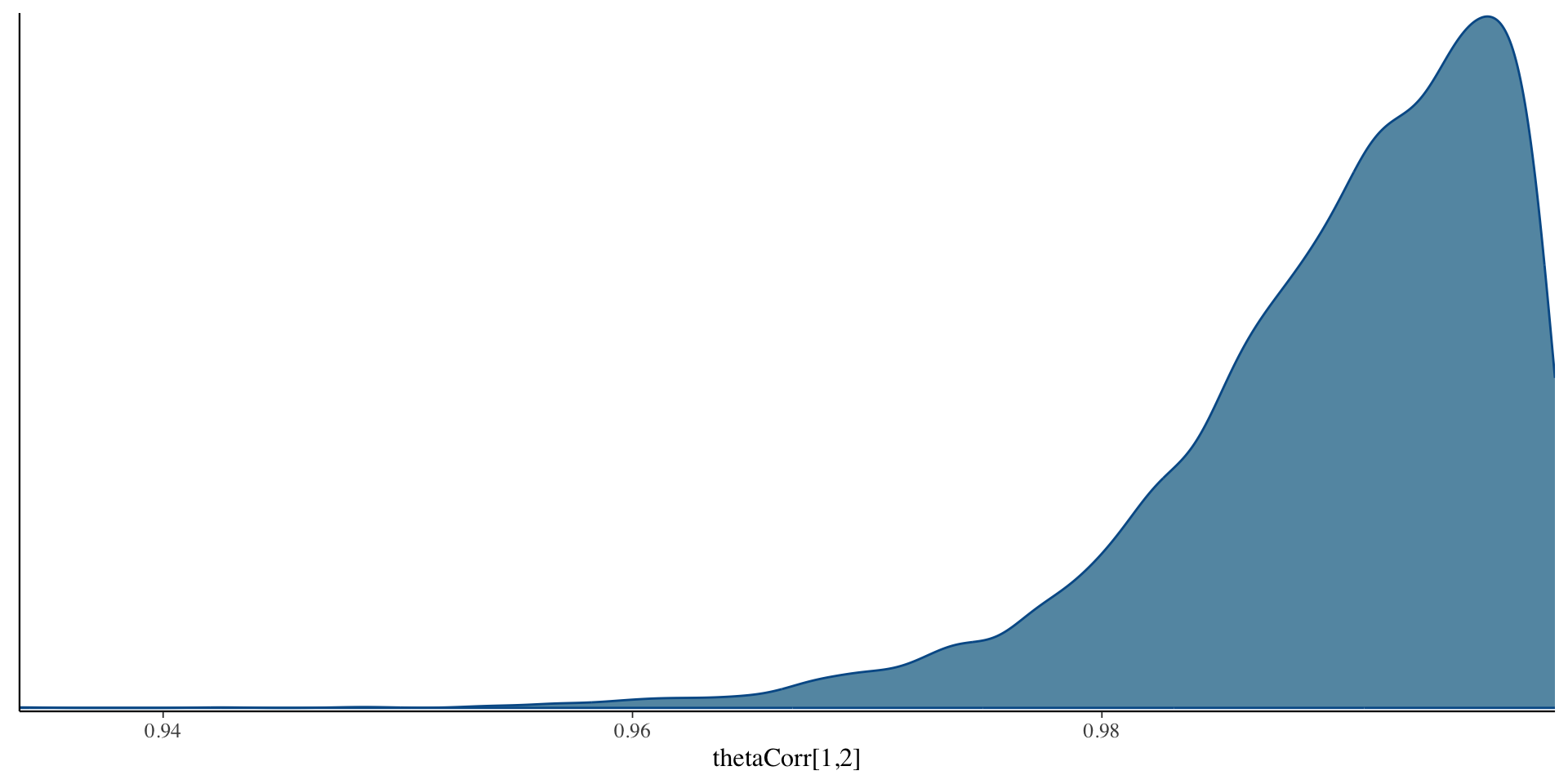
- EAP estimate of correlation was 0.993 – indicates only one factor in data
Correlation Chains
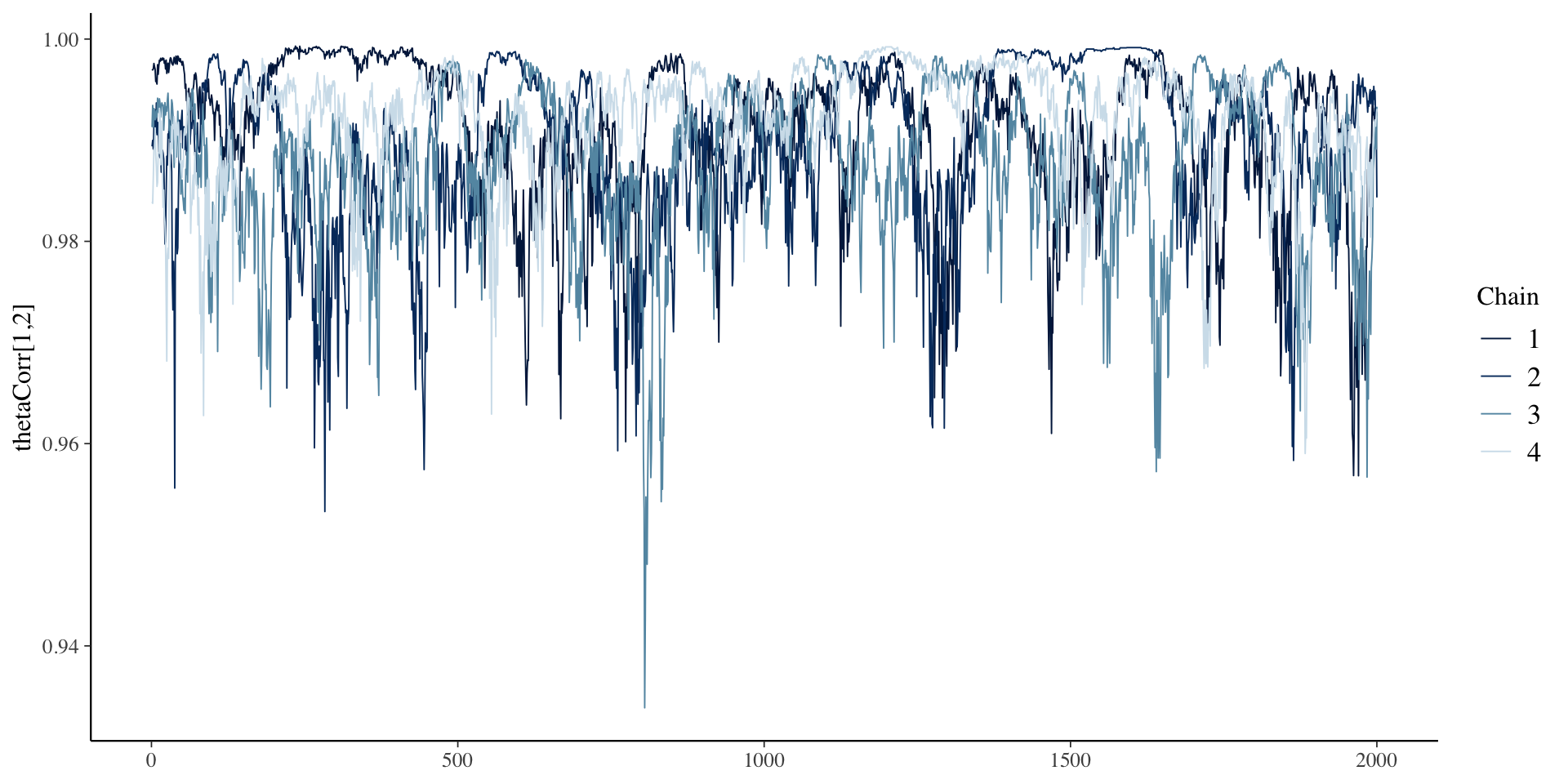
Correlation Posterior Distribution
Example Theta Posterior Distribution
Plots of draws for person 1:
theta[1,1] theta[1,2]
theta[1,1] 1.0000000 0.8221942
theta[1,2] 0.8221942 1.0000000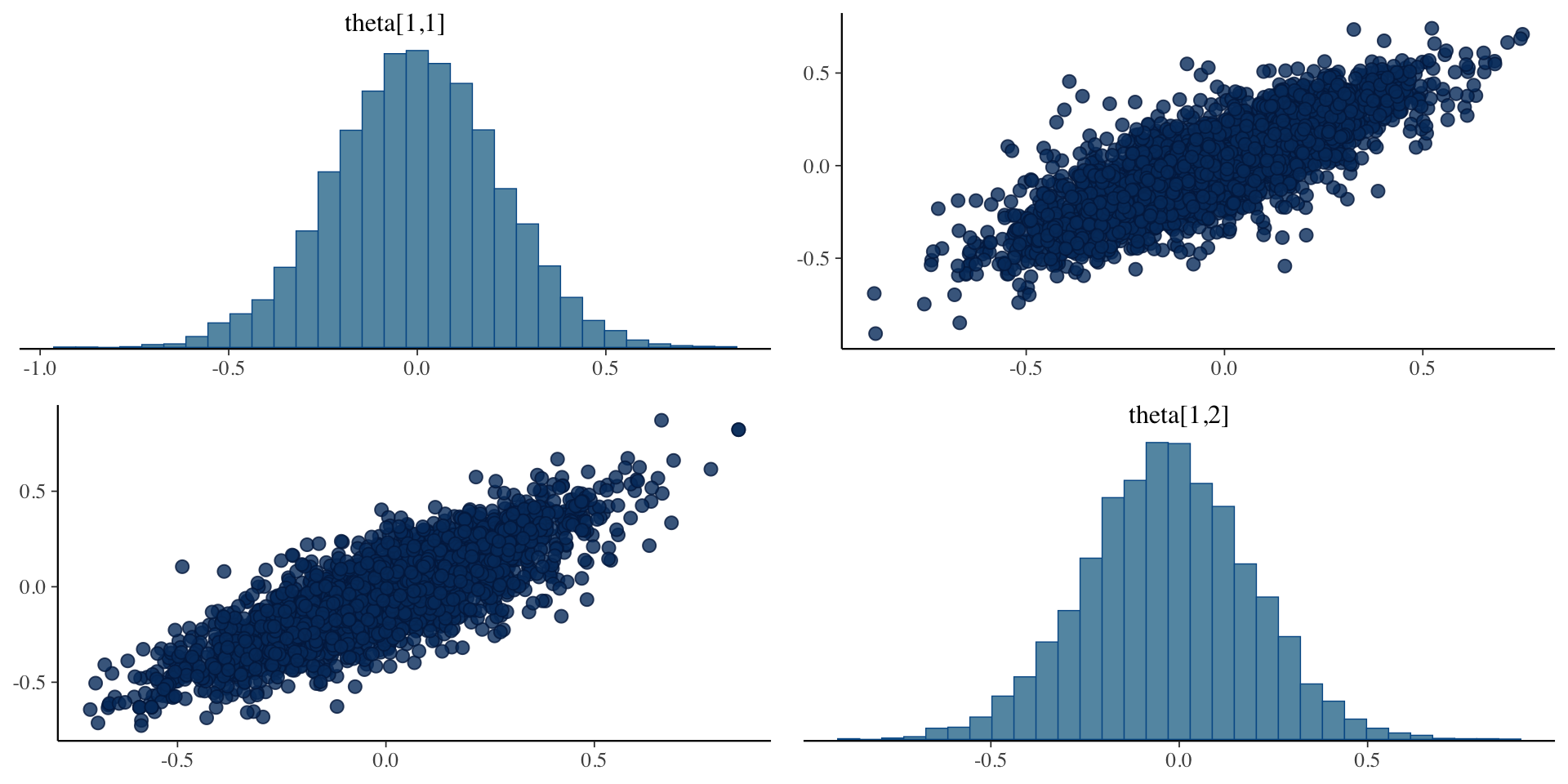
EAP Theta Estimates
Plots of draws for person 1:
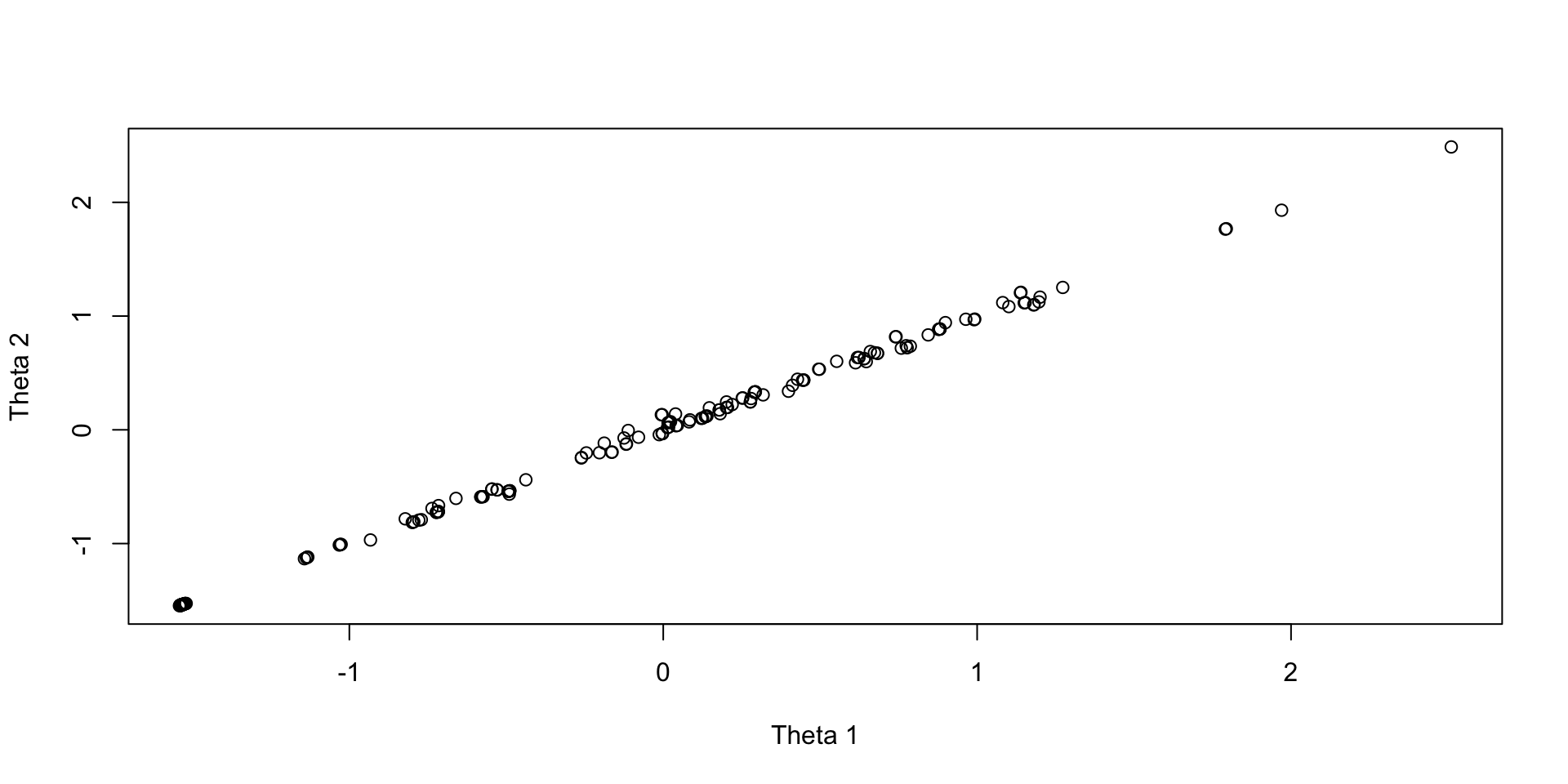
Wrapping Up
Wrapping Up
Stan makes multidimensional latent variable models fairly easy to implement
- LKJ prior allows for scale identification with standardized factors
- Can use my syntax and import any type of Q-matrix
But…
- Estimation is slower (correlations take time)