# A tibble: 7 × 10
variable mean median sd mad q5 q95 rhat ess_bulk ess_tail
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 beta[1] 147. 147. 3.12 3.09 142. 152. 1.00 2961. 4081.
2 beta[2] -0.293 -0.294 0.481 0.465 -1.08 0.497 1.00 3222. 4690.
3 beta[3] -23.1 -23.1 4.36 4.35 -30.2 -16.0 1.00 3268. 4369.
4 beta[4] 81.5 81.5 4.20 4.14 74.7 88.4 1.00 3475. 4302.
5 beta[5] 2.37 2.37 0.690 0.671 1.23 3.50 1.00 3541. 4845.
6 beta[6] 3.52 3.52 0.644 0.628 2.46 4.57 1.00 3767. 4432.
7 sigma 8.27 8.15 1.26 1.21 6.45 10.6 1.00 4323. 4850.Bayesian Model Fit and Comparisons
Lecture 3c
Today’s Lecture Objectives
- Bayesian methods for determining how well a model fits the data (absolute fit: Posterior Predictive Model Checks)
- Bayesian methods for determining which model fits better (relative model fit: Widely Available Information Criterion and Leave One Out methods)
Absolute Model Fit: PPMC
Posterior predictive model checking (PPMC) is a Bayesian method for determining if a model fits the data
- Absolute model fit: “Does my model fit my data well?”
- Overall idea: If a model fits the data well, then simulated data based on the model will resemble the observed data
- Ingredients in PPMC:
- Original data
- Typically characterized by some set of statistics (i.e, sample mean, standard deviation, covariance) applied to data
- Data simulated from posterior draws in the Markov Chain
- Summarized by the same set of statistics
- Original data
PPMC Example: Linear Models
Recall our linear model example with the Diet Data:
\[\text{WeightLB}_p = \beta_0 + \beta_1\text{HeightIN}_p + \beta_2 \text{Group2}_p + \beta_3 \text{Group3}_p + \beta_4\text{HeightIN}_p\text{Group2}_p + \]
\[\beta_5\text{HeightIN}_p\text{Group3}_p + e_p\]
We last used matrices to estimate this model, with the following results:
PPMC Process
The PPMC process is as follows
- Select parameters from a single (sampling) iteration of the Markov chain
- Using the selected parameters and the model, simulate a data set with the same size (number of observations/variables)
- From the simulated data set, calculate selected summary statistics (e.g. mean)
- Repeat steps 1-3 for a fixed number of iterations (perhaps across the whole chain)
- When done, compare position of observed summary statistics to that of the distribution of summary statitsics from simulated data sets (predictive distribution)
Example PPMC
For our model, we have one observed variable that is in the model (\(Y_p\))
- Note, the observations in \(\textbf{X}\) are not modeled, so we do not examine these
First, let’s assemble the posterior draws:
# here, we use format = "draws_matrix" to remove the draws from the array format they default to
posteriorSample = model06_Samples$draws(variables = c("beta", "sigma"), format = "draws_matrix")
posteriorSample# A draws_matrix: 2000 iterations, 4 chains, and 7 variables
variable
draw beta[1] beta[2] beta[3] beta[4] beta[5] beta[6] sigma
1 146 0.353 -27 79 2.43 2.2 9.3
2 149 0.142 -33 79 2.06 2.4 10.5
3 149 0.109 -27 80 2.35 3.0 8.5
4 150 0.153 -21 80 1.72 3.0 10.5
5 140 -0.031 -17 86 1.99 3.1 9.3
6 141 0.307 -19 89 2.46 2.5 9.2
7 149 0.590 -23 79 0.66 2.9 8.0
8 146 -0.362 -22 82 3.02 3.4 7.3
9 146 -0.238 -25 84 2.05 3.6 6.6
10 145 -0.809 -20 81 3.07 3.7 6.7
# ... with 7990 more drawsExample PPMC
Next, we take one draw at random:
Example PPMC
We then generate data based on this sampled iteration and our model distributional assumptions:
[,1]
[1,] 148.273000
[2,] -0.263068
[3,] -27.127900
[4,] 78.629600
[5,] 2.867040
[6,] 3.578690Next, let’s take the mean and standard deviation of the simulated data:
Looping Across All Posterior Samples
We then repeat this process for a set number of samples (here, we’ll use each posterior draw)
simMean = rep(NA, nrow(posteriorSample))
simSD = rep(NA, nrow(posteriorSample))
for (iteration in 1:nrow(posteriorSample)){
betaVector = matrix(data = posteriorSample[iteration, 1:6], ncol = 1)
sigma = posteriorSample[iteration, 7]
conditionalMeans = model06_predictorMatrix %*% betaVector
simData = rnorm(n = N, mean = conditionalMeans, sd = sigma)
simMean[iteration] = mean(simData)
simSD[iteration] = sd(simData)
}
par(mfrow = c(1,2))
hist(simMean)
hist(simSD)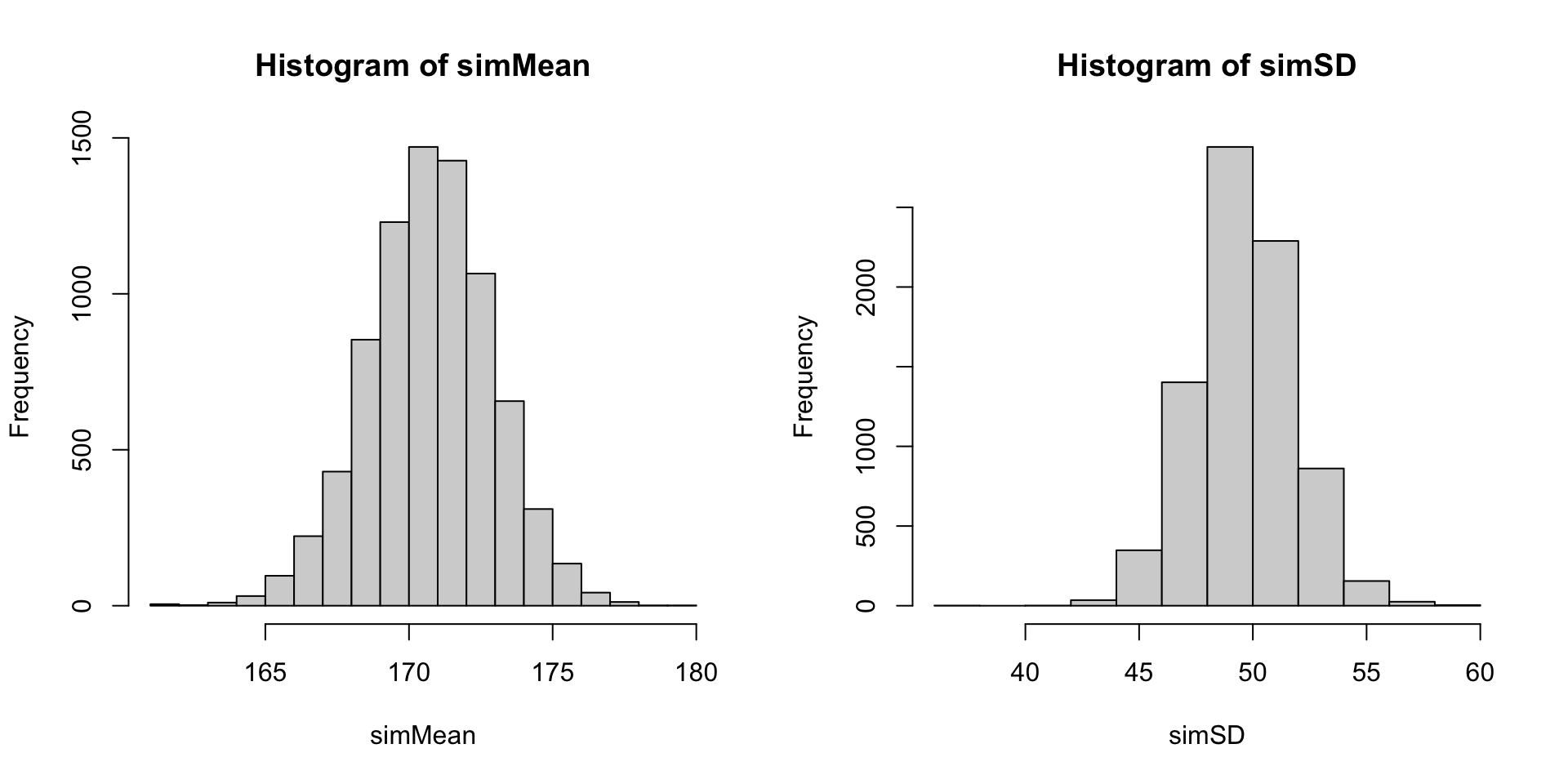
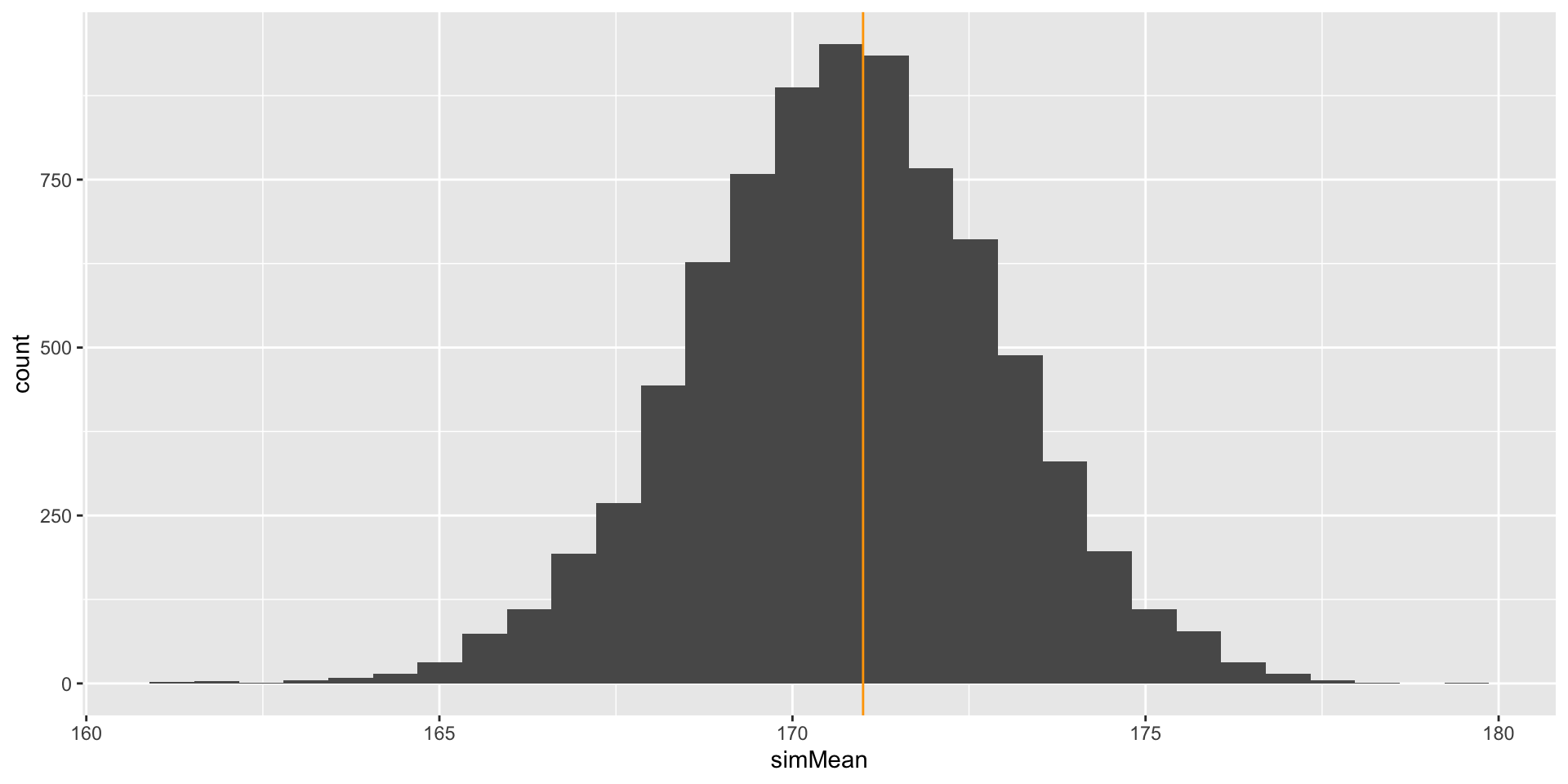
##Comparison with Observed Mean
We can now compare our observed mean and standard deviation with that of the sampled values
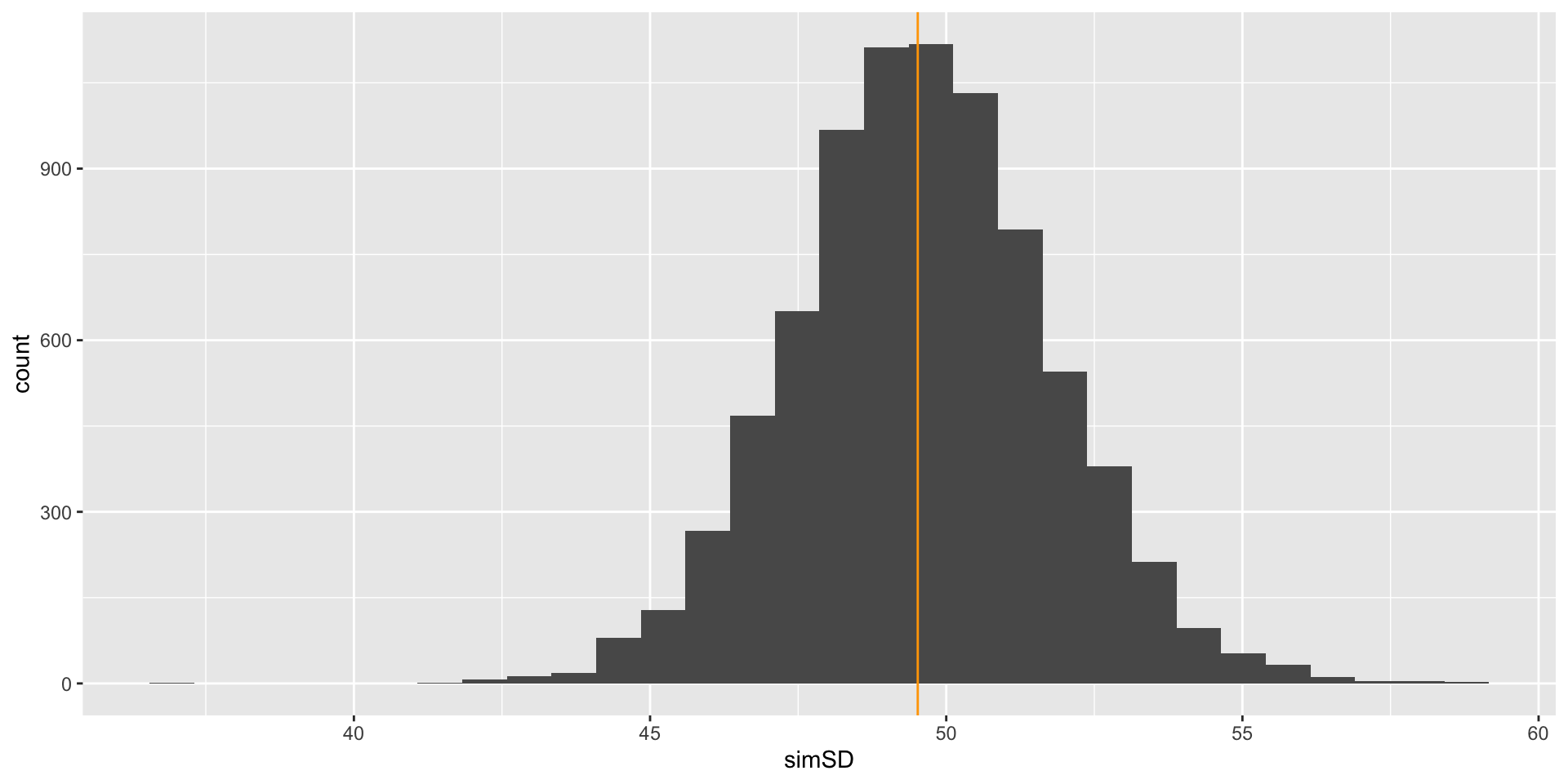
Comparison with Observed SD
We can now compare our observed mean and standard deviation with that of the sampled values
PPMC Charactaristics
PPMC methods are very useful
- They provide a visual way to determine if the model fits the observed data
- They are the main method of assessing absolute fit in Bayesian models
- Absolute fit assesses if a model fits the data
But, there are some drawbacks to PPMC methods
- Almost any statistic can be used
- Some are better than others
- No standard determining how much misfit is too much
- May be overwhelming to compute depending on your model
Posterior Predictive P-Values
We can quantify misfit from PPMC using a type of “p-value”
- The Posterior Predictive P-Value: The proportion of times the statistic from the simulated data exceeds that of the real data
- Useful to determine how far off a statistic is from its posterior predictive distribution
For the mean:
For the standard deviation:
If these p-values were either (a) near zero or (b) near one then this indicates how far off your data are from their predictive distribution
See the example file for comparing between two sets of priors
PPMC and PPP in Stan
We can use the generated quantities section of Stan syntax to compute these for us:
generated quantities{
// general quantities used below:
vector[N] y_pred;
y_pred = X*beta; // predicted value (conditional mean)
// posterior predictive model checking
array[N] real y_rep;
y_rep = normal_rng(y_pred, sigma);
real mean_y = mean(y);
real sd_y = sd(y);
real mean_y_rep = mean(to_vector(y_rep));
real<lower=0> sd_y_rep = sd(to_vector(y_rep));
int<lower=0, upper=1> mean_gte = (mean_y_rep >= mean_y);
int<lower=0, upper=1> sd_gte = (sd_y_rep >= sd(y));
}Which gives us:
# A tibble: 4 × 10
variable mean median sd mad q5 q95 rhat ess_bulk ess_tail
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 mean_y_rep 171. 171. 2.17 2.06 167. 174. 1.00 8737. 7374.
2 sd_y_rep 49.6 49.5 2.20 2.14 46.0 53.2 1.00 8247. 6905.
3 mean_gte 0.441 0 0.496 0 0 1 1.00 8409. NA
4 sd_gte 0.505 1 0.500 0 0 1 1.00 8759. NA Relative Model Fit
Relative Model Fit
Relative model fit: Used to compare two (or more) competing models
- In non-Bayesian models, Information Criteria are often used to make comparisons
- AIC, BIC, etc.
- The model with the lowest index is the model that fits best
- Bayesian model fit is similar
- Uses an index value
- The model with the lowest index is the model that fits best
- Recent advances in Bayesian model fit use indices that are tied to making cross-validation predictions:
- Fit model leaving one observation out
- Calculate statistics related to prediction (for instance, log-likelihood of that observation conditional on model parameters)
- Do for all observations
- Newer Bayesian indices try to mirror these leave-one-out predictions (but approximate these due to time constraints)
Bayesian Model Fit Indices
Way back when (late 1990s and early 2000s), the Deviance Information Criterion was used for relative Bayesian model fit comparisons
\[\text{DIC} = p_D + \overline{D\left(\theta\right)},\]
where the estimated number of parameters is:
\[p_D = \overline{D\left(\theta\right)} - D\left(\bar{\theta}\right), \]
and where
\[ D\left( \theta\right) = -2 \log \left(p\left(y|\theta\right)\right)+C\]
C is a constant that cancels out when model comparisons are made
Here,
- \(\overline{D\left(\theta\right)}\) is the average log likelihood of the data (\(y\)) given the parameters (\(\theta\)) computed across all samples
- \(D\left(\bar{\theta}\right)\) is the log likelihood of the data (\(y\)) computed at the average of the parameters (\(\bar{\theta}\)) computed across all samples
Newer Methods
The DIC has fallen out of favor recently
- Has issues when parameters are discrete
- Not fully Bayesian (point estimate of average of parameter values)
- Can give negative values for estimated numbers of parameters in a model
WAIC (Widely applicable or Watanabe-Akaike information criterion, Watanabe, 2010) corrects some of the problems with DIC:
- Fully Bayesian (uses entire posterior distribution)
- Asymptotically equal to Bayesian cross-validation
- Invariant to parameterization
LOO: Leave One Out via Pareto Smoothed Importance Sampling
More recently, approximations to LOO have gained popularity
- LOO via Pareto Smoothed Importance Sampling attempts to approximate the process of leave-one-out cross-validation using a sampling based-approach
- Gives a finite-sample approximation
- Implemented in Stan
- Can quickly compare models
- Gives warnings when it may be less reliable to use
- The details are very technical, but are nicely compiled in Vehtari, Gelman, and Gabry (2017) (see preprint on arXiv at https://arxiv.org/pdf/1507.04544.pdf)
- Big picture:
- Can compute WAIC and/or LOO via Stan’s
loopackage - LOO also gives variability for model comparisons
- Can compute WAIC and/or LOO via Stan’s
Model Comparisons via Stan
The core of WAIC and LOO is the log-likelihood of the data conditional on the model parameters, as calculated for each observation in the sample of the model:
- We can calculate this using the generated quantities block in Stan:
WAIC in Stan
- Using the
loopackage, we can calculate WAIC with thewaic()function
Computed from 8000 by 30 log-likelihood matrix
Estimate SE
elpd_waic -109.7 6.9
p_waic 6.8 2.7
waic 219.3 13.7
4 (13.3%) p_waic estimates greater than 0.4. We recommend trying loo instead. - Here:
elpd_waicis the expected log pointwise predictive density for WAICp_waicis the WAIC calculation of number of model parameters (a penalty to the likelihood for more parameters)waicis the WAIC index used for model comparisons (lowest value is best fitting; -2*elpd_waic)
Note: WAIC needs a “log_lik” variable in the model analysis to be calculated correctly * That is up to you to provide!!
LOO in Stan
- Using the
loopackage, we can calculate the PSIS-LOO usingcmdstanrobjects with the$loo()function:
Computed from 8000 by 30 log-likelihood matrix
Estimate SE
elpd_loo -110.3 7.2
p_loo 7.5 3.1
looic 220.6 14.4
------
Monte Carlo SE of elpd_loo is NA.
Pareto k diagnostic values:
Count Pct. Min. n_eff
(-Inf, 0.5] (good) 26 86.7% 1476
(0.5, 0.7] (ok) 3 10.0% 433
(0.7, 1] (bad) 1 3.3% 33
(1, Inf) (very bad) 0 0.0% <NA>
See help('pareto-k-diagnostic') for details.Note: LOO needs a “log_lik” variable in the model analysis to be calculated correctly
- That is up to you to provide!!
- If named “log_lik” then you don’t need to provide the function an argument
Notes about LOO
Computed from 8000 by 30 log-likelihood matrix
Estimate SE
elpd_loo -110.3 7.2
p_loo 7.5 3.1
looic 220.6 14.4
------
Monte Carlo SE of elpd_loo is NA.
Pareto k diagnostic values:
Count Pct. Min. n_eff
(-Inf, 0.5] (good) 26 86.7% 1476
(0.5, 0.7] (ok) 3 10.0% 433
(0.7, 1] (bad) 1 3.3% 33
(1, Inf) (very bad) 0 0.0% <NA>
See help('pareto-k-diagnostic') for details.Here:
elpd_loois the expected log pointwise predictive density for LOOp_loois the LOO calculation of number of model parameters (a penalty to the likelihood for more parameters)looicis the LOO index used for model comparisons (lowest value is best fitting; -2*elpd_loo)
Also, you get a warning if to many of your sampled values have bad diagnostic values
Comparing Models with WAIC
To compare models with WAIC, take the one with the lowest value:
Computed from 8000 by 30 log-likelihood matrix
Estimate SE
elpd_waic -109.7 6.9
p_waic 6.8 2.7
waic 219.3 13.7
4 (13.3%) p_waic estimates greater than 0.4. We recommend trying loo instead.
Computed from 8000 by 30 log-likelihood matrix
Estimate SE
elpd_waic -370.2 30.9
p_waic 17.2 4.1
waic 740.3 61.8
12 (40.0%) p_waic estimates greater than 0.4. We recommend trying loo instead. Comparing Models with LOO
To compare models with LOO, the loo package has a built-in comparison function:
# comparing two models with loo:
loo_compare(list(uniformative = model06_Samples$loo(), informative = model06b_Samples$loo())) elpd_diff se_diff
uniformative 0.0 0.0
informative -259.8 33.1 This function calculates the standard error of the difference in ELPD between models
- The SE gives an indication of the standard error in the estimate (relative to the size)
- Can use this to downweight the choice of models when the standard error is high
- Note:
elpd_diffislooicdivided by -2 (on the log-likelihood scale, not the deviance scale)- Here, we interpret the result as the model with uninformative priors is preferred to the model with informative priors
- The size of the elpd_diff is much larger than the standard error, indicating we can be fairly certain of this result
General Points about Bayesian Model Comparison
- Note, WAIC and LOO will converge as sample size increases (WAIC is asymptotic value of LOO)
- Latent variable models present challenges
- Need log likelihood with latent variable integrated out
- Missing data models present challenges
- Need log likelihood with missing data integrated out
- Generally, using LOO is recommended (but providing both is appropriate)
Wrapping Up
The three-part lecture (plus example) using linear models was built to show nearly all parts needed in a Bayesian analysis
- MCMC specifications
- Prior specifications
- Assessing MCMC convergence
- Reporting MCMC results
- Determining if a model fits the data (absolute fit)
- Determining which model fits the data better (relative fit)
All of these topics will be with us when we start psychometric models in our next lecture