IQ perfC
1 78 9
2 84 13
3 84 10
4 85 8
5 87 7
6 91 7
7 92 9
8 94 9
9 94 11
10 96 7
11 99 7
12 105 10
13 105 11
14 106 15
15 108 10
16 112 10
17 113 12
18 115 14
19 118 16
20 134 12Introduction to Missing Data Methods Class
Learning Objectives
- Understand the challenges and implications of missing data in research
- Classify missing data by patterns and mechanisms using Rubin’s framework
- Recognize the limitations of outdated missing data methods
- Explore the design and application of planned missing data methods
Importance of Missing Data
Why Missing Data Matters
- Missing data is pervasive across disciplines (e.g., education, psychology, medicine, political science)
- One big example: Polling errors in elections in 2016/2020 seemed to be affected by missing data (MNAR) - Remedies have been subjective at best
- Mishandling missing data can:
- Bias results –> Inaccurate conclusions
- Reduce statistical power
Modern Methods
- Maximum Likelihood (ML): Estimates parameters directly from observed data likelihood
- Bayesian Estimation: Combines prior beliefs with data likelihood
- Multiple Imputation (MI): Reflects uncertainty by filling in missing data with plausible values
Additional note: Methods here typically require full-information analyses (i.e., likelihoods based on the data directly)
Missing Data Patterns vs. Mechanisms
- A missing data pattern refers to the configuration of observed and missing values in a data set
- What you observe in data
- A missing data mechanism refers to processes that describe different ways in which the probability of missing values relates to the data
- Typically untestable
- What is assumed about data
- Patterns describe where the holes are in the data, whereas mech- anisms describe why the values are missing
Missing Data Patterns
Types of Missing Data Patterns
- Univariate
- Monotone
- General
- Planned Missingness
- Latent Variable
- Underidentified
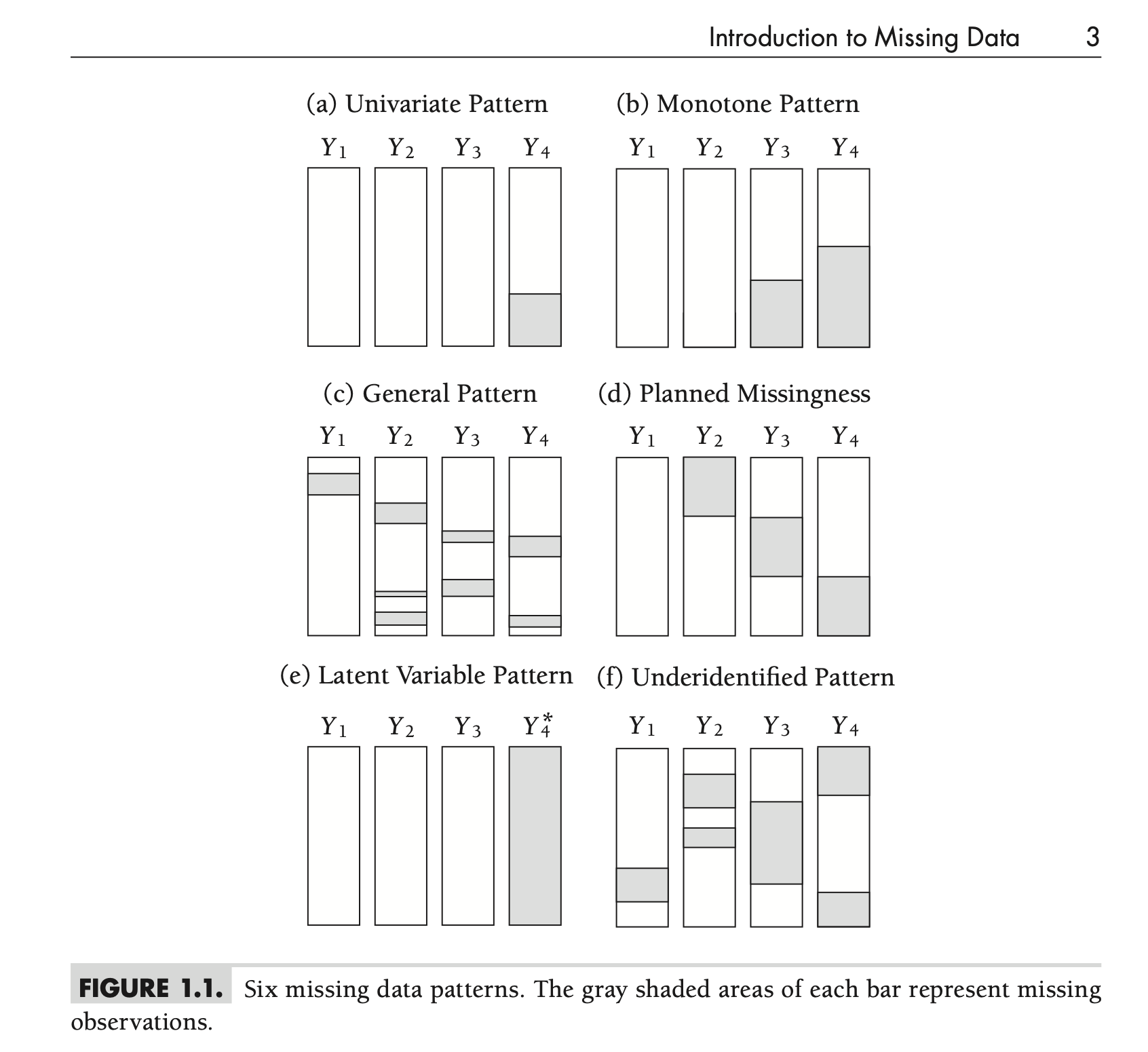
Univariate Pattern
- Missing values restricted to one variable
- Example: Missing outcomes for some participants
Monotone Pattern
- Missing data accumulates predictably
- Example: Dropout in longitudinal studies
- Can be treated without complicated iterative estimation algorithms
General Pattern
- Missing data scattered randomly across the dataset
- The three contemporary methods (maximum likelihood, Bayesian estimation, and multiple imputation) work well with this configuration
- Generally no reason to choose an analytic method based on the missing data pattern alone
Planned Missingness
- Variables are intentionally missing for a large proportion of respondents
- Can reduce respondent burden and research costs
- Often with minimal impact on statistical power
Latent Variable Pattern
- Latent variables are essentially missing data
- Presents challenges in secondary analyses
- Example: Iowa wishes to understand how well an incoming student’s ACT score predicts first year GPA
- ACT Score: An estimate–not an observation
- Can think of scores as single imputation
- ACT Score: An estimate–not an observation
Underidentified Pattern
- Insufficient overlap of data for estimation
- Example: Sparse cell counts for categorical variables
Missing Data Mechanisms
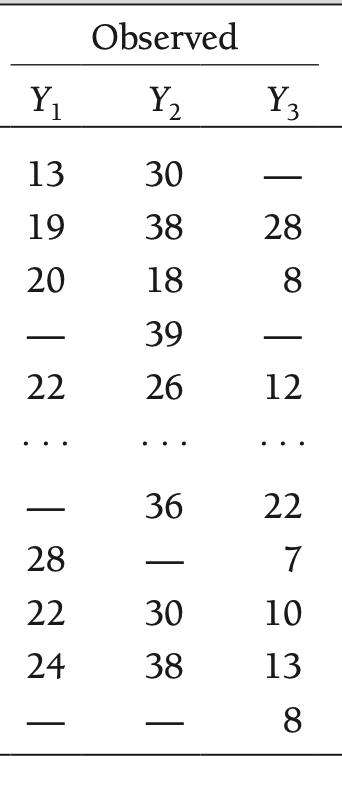
Hypothetical Data Partitioning: Observed Data
- Before getting to the types of missing data mechanisms, we must first define some notation
- Our observed data matrix will be defined as \(\mathbf{Y}_{(\text{obs})}\)
- Here, \(\mathbf{Y}_{(\text{obs})} = \left[Y_1, Y_2, Y_3 \right]\)
Hypothetical Data Partitioning: Complete Data
- Imagine if you could somehow see what the values of the missing data were – the complete data
- Our hypothetical data matrix will be defined as \(\mathbf{Y}_{(\text{com})}\) (sometimes denoted \(\mathbf{Y}_{(1)}\))
- Note: This is not possible through any method and is only a hypothetical example to help define missing data mechanisms

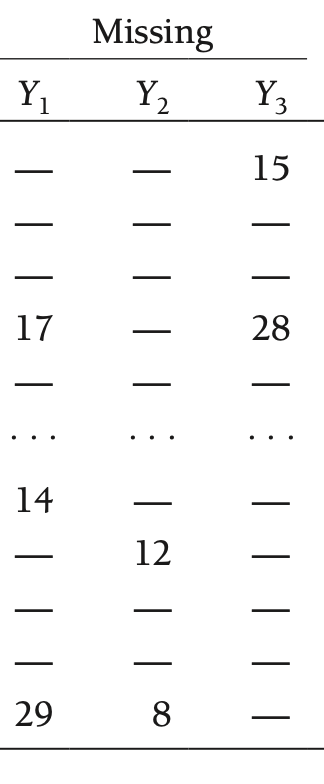
Hypothetical Data Partitioning: Missing Data
- Now, take the values that were missing and only create a matrix of those terms
- Our hypothetical data matrix will be defined as \(\mathbf{Y}_{(\text{mis})}\) (sometimes denoted \(\mathbf{Y}_{(0)}\))
- Note: Again, this is not possible through any method and is only a hypothetical example to help define missing data mechanisms
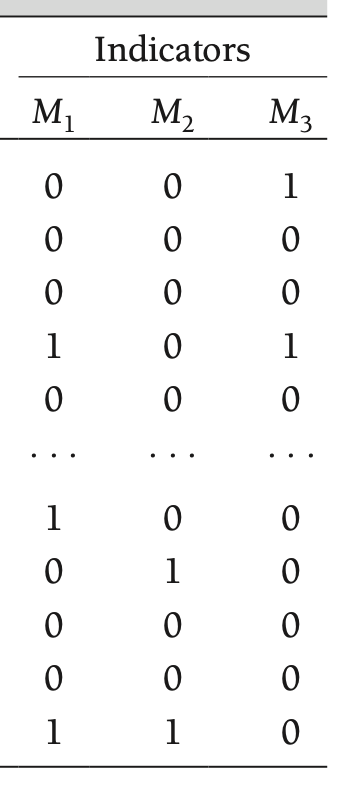
Rubin’s Framework
- Models that explain whether a participant has missing values
- How those tendencies relate to the realized data in \(\mathbf{Y}_{(\text{obs})}\) or \(\mathbf{Y}_{(\text{mis})}\)
- Here, \(\mathbf{M} = \left[M_1, M_2, M_3 \right]\)
Example Data
- To demonstrate some of the ideas of types of missing data, let’s consider a situation where you have collected two variables:
- IQ scores
- Job performance
- Imagine you are an employer looking to hire employees for a job where IQ is important
Missing Data Mechanisms
Missing Data Mechanisms
A very rough typology of missing data puts missing observations into three categories:
- Missing Completely At Random (MCAR)
- Missing At Random (MAR)
- Missing Not At Random (MNAR)
Missing Completely At Random (MCAR)
- Missing data are MCAR if the events that lead to missingness are independent of:
- The observed variables
- -and-
- The unobserved parameters of interest
- Examples:
- Planned missingness in survey research
- Some large-scale tests are sampled using booklets
- Students receive only a few of the total number of items
- The items not received are treated as missing – but that is completely a function of sampling and no other mechanism
- Planned missingness in survey research
A Formal MCAR Definition
Formally, we note that data are MCAR if the probability of the data being missing is independent of the observed data \(\mathbf{Y}_{(obs)}\) and the missing data \(\mathbf{Y}_{(mis)}\):
\[Pr \left(\mathbf{M} = 1 \mid \mathbf{Y}_{(obs)}, \mathbf{Y}_{(mis)}, \boldsymbol{\phi} \right) = Pr \left(\mathbf{M} = 1 \mid \boldsymbol{\phi} \right)\]
- Here, \(\boldsymbol{\phi}\) are model parameters that define the overall probabilities of missing data
- Like saying a missing observation is due to pure randomness (such as missing if a coin flipped falls on heads)
Implications of MCAR
- Because the mechanism of missing is not due to anything other than chance, inclusion of MCAR in data will not bias your results
- Can use methods based on listwise deletion, multiple imputation, or maximum likelihood
- Your effective sample size is lowered, though
- Less power, less efficiency
MCAR Data
Missing data are dispersed randomly throughout data
IQ perfMCAR
1 78 NA
2 84 13
3 84 NA
4 85 8
5 87 7
6 91 7
7 92 9
8 94 9
9 94 11
10 96 NA
11 99 7
12 105 10
13 105 11
14 106 15
15 108 10
16 112 NA
17 113 12
18 115 14
19 118 16
20 134 NAMCAR vs. Complete Data Comparison
Complete Data
lavaan 0.6-19 ended normally after 20 iterations
Estimator ML
Optimization method NLMINB
Number of model parameters 5
Number of observations 20
Model Test User Model:
Test statistic 0.000
Degrees of freedom 0
Parameter Estimates:
Standard errors Standard
Information Expected
Information saturated (h1) model Structured
Covariances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
IQ ~~
perfC 19.500 9.151 2.131 0.033 19.500 0.542
Intercepts:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
IQ 100.000 3.079 32.478 0.000 100.000 7.262
perfC 10.350 0.584 17.714 0.000 10.350 3.961
Variances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
IQ 189.600 59.957 3.162 0.002 189.600 1.000
perfC 6.828 2.159 3.162 0.002 6.828 1.000MAR Data
lavaan 0.6-19 ended normally after 20 iterations
Estimator ML
Optimization method NLMINB
Number of model parameters 5
Number of observations 15
Model Test User Model:
Test statistic 0.000
Degrees of freedom 0
Parameter Estimates:
Standard errors Standard
Information Expected
Information saturated (h1) model Structured
Covariances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
IQ ~~
perfMCAR 19.360 9.299 2.082 0.037 19.360 0.638
Intercepts:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
IQ 99.733 2.777 35.916 0.000 99.733 9.274
perfMCAR 10.600 0.729 14.539 0.000 10.600 3.754
Variances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
IQ 115.662 42.234 2.739 0.006 115.662 1.000
perfMCAR 7.973 2.911 2.739 0.006 7.973 1.000Missing at Random Definition
Formally, we note that data are MAR if the probability of the data being missing is related to the observed data \(\mathbf{Y}_{(obs)}\) but not the missing data \(\mathbf{Y}_{(mis)}\):
\[Pr \left(\mathbf{M} = 1 \mid \mathbf{Y}_{(obs)}, \mathbf{Y}_{(mis)}, \boldsymbol{\phi} \right) = Pr \left(\mathbf{M} = 1 \mid \mathbf{Y}_{(obs)}, \boldsymbol{\phi} \right)\]
- Again, \(\boldsymbol{\phi}\) are model parameters that define the overall probabilities of missing data
- Like saying a missing observation is due to pure randomness (such as missing if a coin flipped falls on heads)
MAR Data
Missing data are related to other data:
- Any IQ less than 90 did not have a performance variable
- Could be that anyone with an IQ of 90 or less was not hired
- Not hired means not having job performance data
IQ perfMAR
1 78 NA
2 84 NA
3 84 NA
4 85 NA
5 87 NA
6 91 7
7 92 9
8 94 9
9 94 11
10 96 7
11 99 7
12 105 10
13 105 11
14 106 15
15 108 10
16 112 10
17 113 12
18 115 14
19 118 16
20 134 12Implications of MAR
- If data are missing at random, biased results could occur
- Inferences based on listwise deletion will be biased and inefficient
- Fewer data points = more error in analysis
- Inferences based on maximum likelihood will be unbiased but inefficient
- The first eight chapters of the book focus on methods for MAR data
MAR vs. Complete Data Comparison
Complete Data
lavaan 0.6-19 ended normally after 20 iterations
Estimator ML
Optimization method NLMINB
Number of model parameters 5
Number of observations 20
Model Test User Model:
Test statistic 0.000
Degrees of freedom 0
Parameter Estimates:
Standard errors Standard
Information Expected
Information saturated (h1) model Structured
Covariances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
IQ ~~
perfC 19.500 9.151 2.131 0.033 19.500 0.542
Intercepts:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
IQ 100.000 3.079 32.478 0.000 100.000 7.262
perfC 10.350 0.584 17.714 0.000 10.350 3.961
Variances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
IQ 189.600 59.957 3.162 0.002 189.600 1.000
perfC 6.828 2.159 3.162 0.002 6.828 1.000MAR Data
lavaan 0.6-19 ended normally after 21 iterations
Estimator ML
Optimization method NLMINB
Number of model parameters 5
Number of observations 15
Model Test User Model:
Test statistic 0.000
Degrees of freedom 0
Parameter Estimates:
Standard errors Standard
Information Expected
Information saturated (h1) model Structured
Covariances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
IQ ~~
perfMAR 19.489 9.413 2.070 0.038 19.489 0.633
Intercepts:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
IQ 105.467 2.947 35.791 0.000 105.467 9.241
perfMAR 10.667 0.697 15.302 0.000 10.667 3.951
Variances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
IQ 130.249 47.560 2.739 0.006 130.249 1.000
perfMAR 7.289 2.662 2.739 0.006 7.289 1.000Missing Not At Random (MNAR) Definition
Formally, we note that data are MNAR if the probability of the data being missing is related to the observed data \(\mathbf{Y}_{(obs)}\) and the missing data \(\mathbf{Y}_{(mis)}\):
\[Pr \left(\mathbf{M} = 1 \mid \mathbf{Y}_{(obs)}, \mathbf{Y}_{(mis)}, \boldsymbol{\phi} \right)\]
- Again, \(\boldsymbol{\phi}\) are model parameters that define the overall probabilities of missing data
Often called non-ignorable missingness
- Inferences based on listwise deletion or maximum likelihood will be biased and inefficient
- Need to provide statistical model for missing data simultaneously with estimation of original model
Surviving Missing data: A Brief Guide
Using Statistical Methods with Missing Data
- Missing data can alter your analysis results dramatically depending upon:
- 1. The type of missing data
- 2. The type of analysis algorithm
- The choice of an algorithm and missing data method is important in avoiding issues due to missing data
The Worst Case Scenario: MNAR
- The worst case scenario is when data are MNAR: missing not at random
- Non-ignorable missing
- You cannot easily get out of this mess
- Instead you have to be clairvoyant
- Analyses algorithms must incorporate models for missing data
- And these models must also be right
The Reality
- In most empirical studies, MNAR as a condition is an afterthought
- It is impossible to know definitively if data truly are MNAR
- So data are treated as MAR or MCAR
- Hypothesis tests do exist for MCAR (i.e., Little’s test)
- But, often this test is rejected
The Best Case Scenario: MCAR
- Under MCAR, pretty much anything you do with your data will give you the “right” (unbiased) estimates of your model parameters
- MCAR is very unlikely to occur
- In practice, MCAR is treated as equally unlikely as MNAR
The Middle Ground: MAR
- MAR is the common compromise used in most empirical research
- Under MAR, maximum likelihood algorithms are unbiased
- Maximum likelihood is for many methods:
- Linear mixed models i
- Models with “latent” random effects (CFA/SEM models)
Outdated Methods for Handling Missing Data
Bad Ways to Handle Missing Data
- Dealing with missing data is important, as the mechanisms you choose can dramatically alter your results
- This point was not fully realized when the first methods for missing data were created
- Each of the methods described in this section should never be used
- Given to show perspective – and to allow you to understand what happens if you were to choose each
Deletion Methods
- Deletion methods are just that: methods that handle missing data by deleting observations
- Listwise deletion: delete the entire observation if any values are missing
- Pairwise deletion: delete a pair of observations if either of the values are missing
- Assumptions: Data are MCAR
- Limitations:
- Reduction in statistical power if MCAR
- Biased estimates if MAR or MNAR
Listwise Deletion
- Listwise deletion discards all of the data from an observation if one or more variables are missing
- Most frequently used in statistical software packages that are not optimizing a likelihood function (need ML)
- In linear models:
- R lm() list-wise deletes cases where DVs are missing
Listwise Deletion Example: MCAR
Parameter Estimates:
Standard errors Standard
Information Expected
Information saturated (h1) model Structured
Regressions:
Estimate Std.Err z-value P(>|z|)
perfC ~
IQ 0.103 0.036 2.884 0.004
Intercepts:
Estimate Std.Err z-value P(>|z|)
.perfC 0.065 3.600 0.018 0.986
Variances:
Estimate Std.Err z-value P(>|z|)
.perfC 4.822 1.525 3.162 0.002
Parameter Estimates:
Standard errors Standard
Information Expected
Information saturated (h1) model Structured
Regressions:
Estimate Std.Err z-value P(>|z|)
perfMCAR ~
IQ 0.167 0.052 3.205 0.001
Intercepts:
Estimate Std.Err z-value P(>|z|)
.perfMCAR -6.094 5.239 -1.163 0.245
Variances:
Estimate Std.Err z-value P(>|z|)
.perfMCAR 4.733 1.728 2.739 0.006Listwise Deletion Example: MAR
Parameter Estimates:
Standard errors Standard
Information Expected
Information saturated (h1) model Structured
Regressions:
Estimate Std.Err z-value P(>|z|)
perfC ~
IQ 0.103 0.036 2.884 0.004
Intercepts:
Estimate Std.Err z-value P(>|z|)
.perfC 0.065 3.600 0.018 0.986
Variances:
Estimate Std.Err z-value P(>|z|)
.perfC 4.822 1.525 3.162 0.002
Parameter Estimates:
Standard errors Standard
Information Expected
Information saturated (h1) model Structured
Regressions:
Estimate Std.Err z-value P(>|z|)
perfMAR ~
IQ 0.150 0.047 3.163 0.002
Intercepts:
Estimate Std.Err z-value P(>|z|)
.perfMAR -5.114 5.019 -1.019 0.308
Variances:
Estimate Std.Err z-value P(>|z|)
.perfMAR 4.373 1.597 2.739 0.006Pairwise Deletion
- Pairwise deletion discards a pair of observations if either one is missing
- Different from listwise: uses more data (rest of data not thrown out)
- Assumes: MCAR
- Limitations:
- Reduction in statistical power if MCAR
- Biased estimates if MAR or MNAR
- Can be an issue when forming covariance/correlation matrices
- May make them non-invertible, problem if used as input into statistical procedures
Pairwise Deletion Example
Single Imputation Methods
- Single imputation methods replace missing data with some type of value
- Single: one value used
- Imputation: replace missing data with value
- Upside: can use entire data set if missing values are replaced
- Downside: biased parameter estimates and standard errors (even if missing is MCAR)
- Type-I error issues
- Still: never use these techniques
Unconditional Mean Imputation
- Unconditional mean imputation replaces the missing values of a variable with its estimated mean
- Unconditional = mean value without any input from other variables
Unconditional Mean Imputation: MCAR Data vs Complete Data
Complete
Call:
lm(formula = perfC ~ IQ, data = jobPerf)
Residuals:
Min 1Q Median 3Q Max
-3.2472 -1.6498 -0.6301 1.2742 4.2956
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.06519 3.79433 0.017 0.9865
IQ 0.10285 0.03759 2.736 0.0136 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.315 on 18 degrees of freedom
Multiple R-squared: 0.2937, Adjusted R-squared: 0.2545
F-statistic: 7.487 on 1 and 18 DF, p-value: 0.01357MCAR
Call:
lm(formula = perfMCAR_meanImpute ~ IQ, data = jobPerf)
Residuals:
Min 1Q Median 3Q Max
-3.5234 -1.2723 -0.4509 1.3402 4.0215
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.94177 3.81241 0.772 0.4503
IQ 0.07658 0.03777 2.028 0.0577 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.326 on 18 degrees of freedom
Multiple R-squared: 0.1859, Adjusted R-squared: 0.1407
F-statistic: 4.112 on 1 and 18 DF, p-value: 0.05766Unconditional Mean Imputation: MAR Data vs Complete Data
Complete
Call:
lm(formula = IQ ~ perfC, data = jobPerf)
Residuals:
Min 1Q Median 3Q Max
-23.569 -7.425 1.216 6.572 29.287
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 70.439 11.143 6.322 5.87e-06 ***
perfC 2.856 1.044 2.736 0.0136 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 12.2 on 18 degrees of freedom
Multiple R-squared: 0.2937, Adjusted R-squared: 0.2545
F-statistic: 7.487 on 1 and 18 DF, p-value: 0.01357MAR
Call:
lm(formula = IQ ~ perfMAR_meanImpute, data = jobPerf)
Residuals:
Min 1Q Median 3Q Max
-22.000 -8.418 2.272 7.288 30.435
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 71.480 13.506 5.293 4.95e-05 ***
perfMAR_meanImpute 2.674 1.237 2.162 0.0443 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 12.93 on 18 degrees of freedom
Multiple R-squared: 0.2061, Adjusted R-squared: 0.162
F-statistic: 4.674 on 1 and 18 DF, p-value: 0.04434Conditional Mean Imputation (Regression)
- Conditional mean imputation uses regression analyses to impute missing values
- The missing values are imputed using the predicted values in each regression (conditional means)
- For our data we would form regressions for each outcome using the other variables
- PERF = β01 + β11*IQ
- More accurate than unconditional mean imputation
- But still provides biased parameters and SEs
Stochastic Conditional Mean Imputation
- Stochastic conditional mean imputation adds a random component to the imputation
- Representing the error term in each regression equation
- Assumes MAR rather than MCAR
- Better than any other of these methods (and the basis for multiple imputation)
Imputation by Proximity: Hot Deck Matching
- Hot deck matching uses real data – from other observations as its basis for imputing
- Observations are “matched” using similar scores on variables in the data set
- Imputed values come directly from matched observations
- Upside: Helps to preserve univariate distributions; gives data in an appropriate range
- Downside: biased estimates (especially of regression coefficients), too-small standard errors
Scale Imputation by Averaging
- In psychometric tests, a common method of imputation has been to use a scale average rather than total score
- Can re-scale to total score by taking # items * average score
- Problem: treating missing items this way is like using person mean
- Reduces standard errors
- Makes calculation of reliability biased
Longitudinal Imputation: Last Observation Carried Forward
- A commonly used imputation method in longitudinal data has been to treat observations that dropped out by carrying forward thelast observation
- More common in medical studies and clinical trials
- Assumes scores do not change after dropout – bad idea
- Thought to be conservative
- Can exaggerate group differences
- Limits standard errors that help detect group differences
Why Single Imputation Is Bad Science
- Overall, the methods described in this section are not useful for handling missing data
- If you use them you will likely get a statistical answer that is an artifact
- Actual estimates you interpret (parameter estimates) will be biased (in either direction)
- Standard errors will be too small
- Leads to Type-I Errors
- Putting this together: you will likely end up making conclusions about your data that are wrong
Auxiliary Variables and Semi-Partial Correlation
Auxiliary Variables and Semi-Partial Correlation
The use of auxiliary variables can help mitigate bias and improve statistical power when missing data are present
- An auxiliary variable is an extraneous variable that correlates with missingness or the outcome but is not part of the main analysis
A key aspect of auxiliary variable selection is understanding their semi-partial correlation with the outcome
- Semi-partial correlation quantifies the unique contribution of a predictor to the outcome, after removing its shared variance with other predictors
- This makes Semi-partial correlations useful for identifying the most informative auxiliary variables
What is a Semi-Partial Correlation?
- Semi-partial correlation (also known as part correlation) measures the unique association between a predictor and the outcome while controlling for other variables in the model
- Unlike partial correlation, which removes shared variance from both variables, semi-partial correlation removes shared variance only from the predictor variable, leaving the outcome variable unchanged
What is a Semi-Partial Correlation?
Mathematically, the semi-partial correlation of a predictor \(( X_1 )\) with an outcome \((Y)\), controlling for another predictor \(X_2\), is given by:
\[ r_{Y(X_1.X_2)} = \frac{r_{YX_1} - r_{YX_2}r_{X_1X_2}}{\sqrt{1 - r_{X_1X_2}^2}} \]
where: - \(r_{YX_1}\)is the correlation between \(Y\) and \(X_1\), - \(r_{YX_2}\) is the correlation between \(Y\) and \(X_2\), - \(r_{X_1X_2}\) is the correlation between \(X_1\) and \(X_2\).
Why is Semi-Partial Correlation Important?
Semi-partial correlation is widely used in statistics for several reasons:
- Understanding Unique Contributions – It helps to determine how much unique variance a predictor explains in the outcome, which is useful in multiple regression and hierarchical modeling
- Feature Selection – In machine learning and predictive modeling, semi-partial correlation can help identify the most important predictors
- Model Interpretation – Researchers can use it to assess the relative importance of predictors in explaining variance in an outcome
- Comparing Predictors – It allows us to compare multiple predictors to see which has the most unique explanatory power
Because semi-partial correlation removes shared variance only from the predictor, it reflects the real-world scenario where some predictors contribute uniquely to an outcome while sharing variance with others.
Semi-Partial Correlation in the Context of Missing Data
- Semi-partial correlations help identify which auxiliary variables provide unique information about missingness or the outcome
- This ensures that missing data handling methods, such as multiple imputation or maximum likelihood estimation, make the most use of available information
Example in R
Simulated Data
Let’s create a dataset where we examine semi-partial correlations:
Computing Semi-Partial Correlation using lm() residuals
Computing Semi-Partial Correlation using the ppcor package
$estimate
y x1 x2 aux
y 1.0000000 -0.2359666 -0.3182904 0.3371851
x1 -0.1543577 1.0000000 -0.3222919 0.7896905
x2 -0.2605867 -0.4033669 1.0000000 0.6280408
aux 0.1904317 0.6817890 0.4332414 1.0000000
$p.value
y x1 x2 aux
y 0.000000000 1.932959e-02 1.403247e-03 6.859535e-04
x1 0.129123301 0.000000e+00 1.210448e-03 4.366625e-22
x2 0.009555429 3.816958e-05 0.000000e+00 4.454870e-12
aux 0.060351647 1.092502e-14 8.354724e-06 0.000000e+00
$statistic
y x1 x2 aux
y 0.000000 -2.379176 -3.289682 3.509232
x1 -1.530736 0.000000 -3.335800 12.611715
x2 -2.644587 -4.319133 0.000000 7.907571
aux 1.900622 9.131503 4.709848 0.000000
$n
[1] 100
$gp
[1] 2
$method
[1] "pearson"Comparison of Results
Both methods should yield similar results, but spcor provides a more automated way to compute semi-partial correlations, making it easier to handle multiple predictors at once.
- But, ‘spcor’ doesn’t have the ability to specify certain combinations of variables
Interpretation:
- If the semi-partial correlation is strong (e.g., above 0.30 in absolute value), the auxiliary variable is likely to contain unique information about the outcome and should be included in the missing data handling process
Wrapping Up
Lecture Summary
- Missing data are common in statistical analyses
- They are frequently neglected
- MNAR: hard to model missing data and observed data simultaneously
- MCAR: doesn’t often happen
- MAR: most missing imputation assumes MVN
- More often than not, ML is the best choice
- Software is getting better at handling missing data
- We will discuss how ML works next week