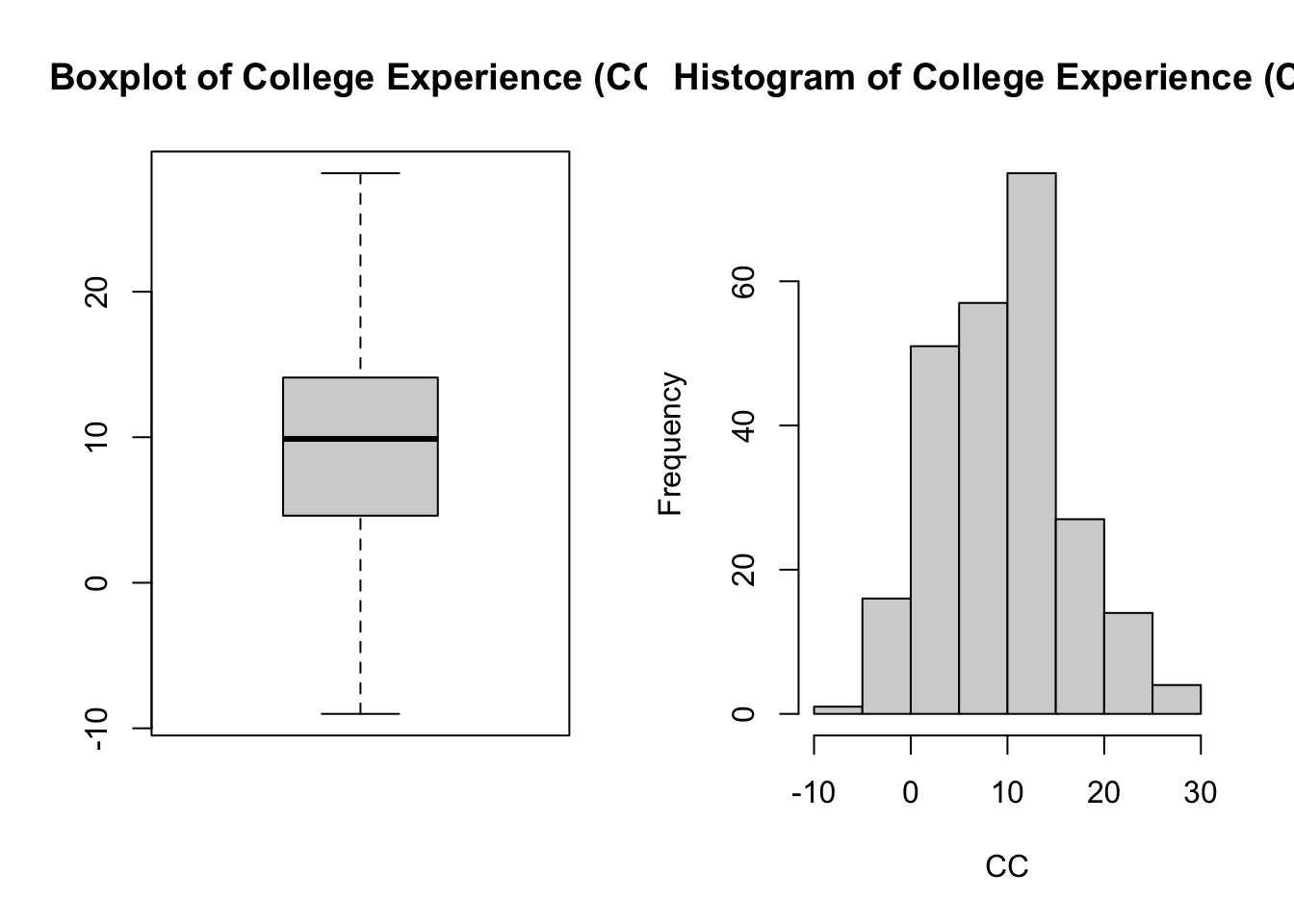
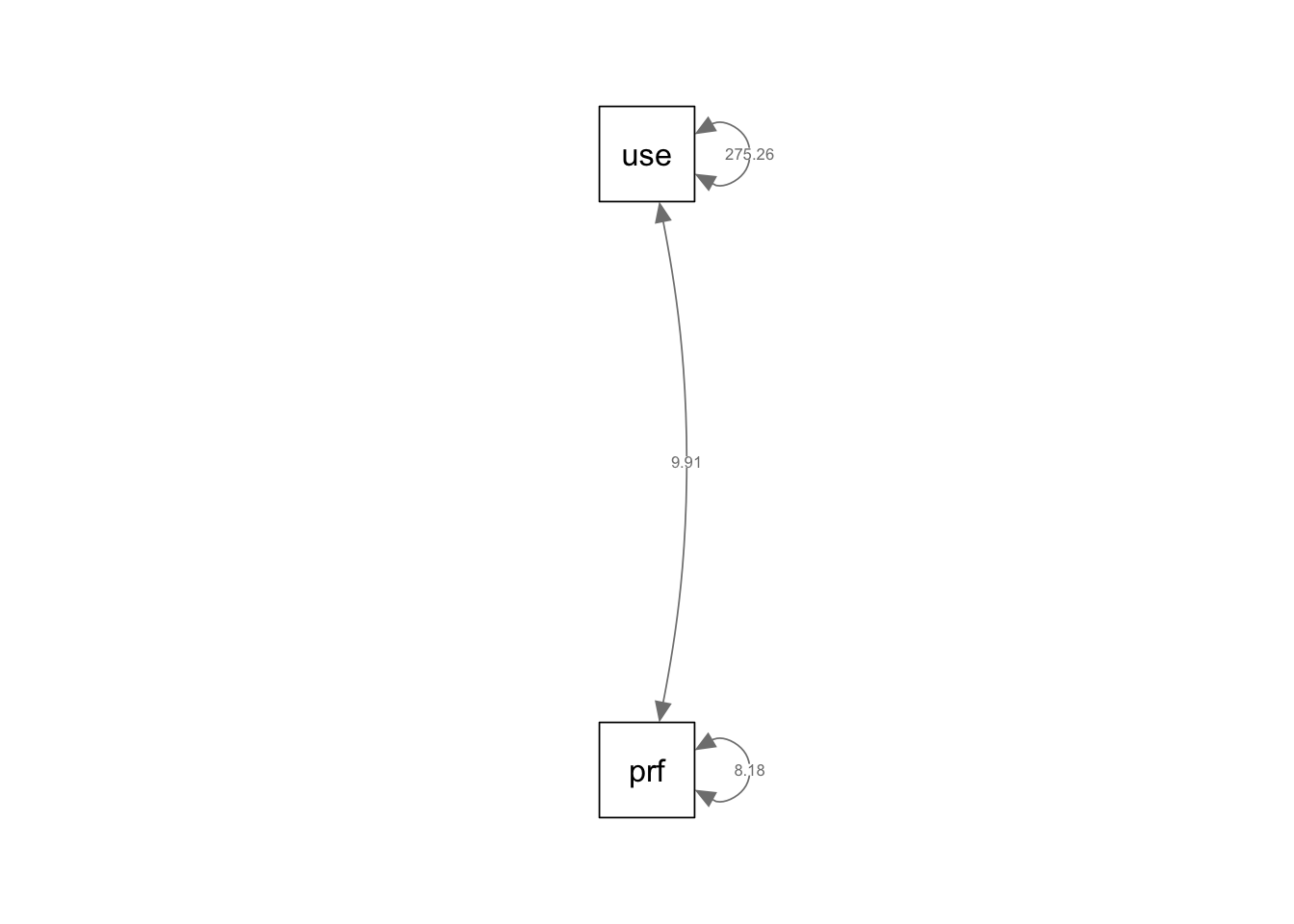
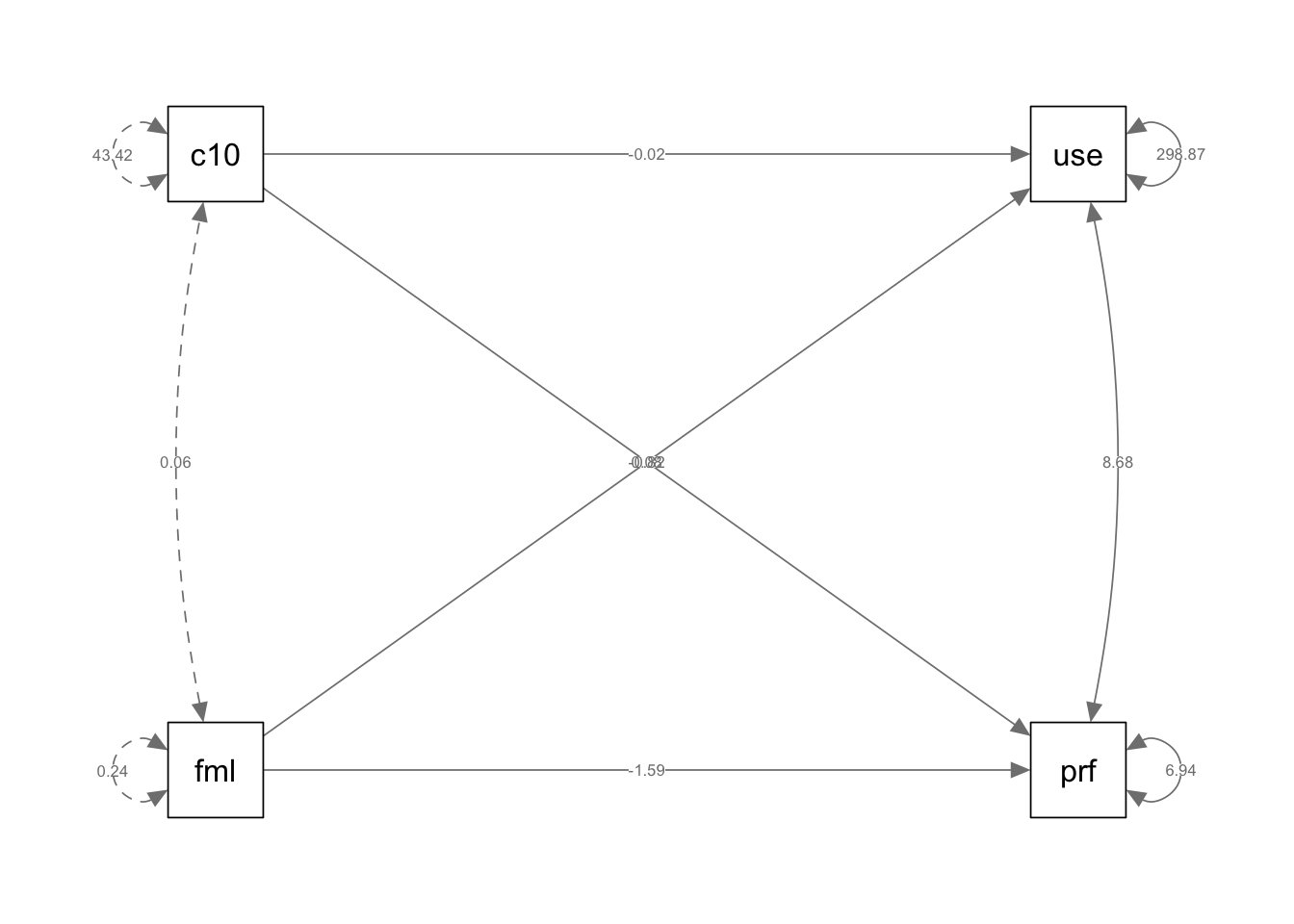
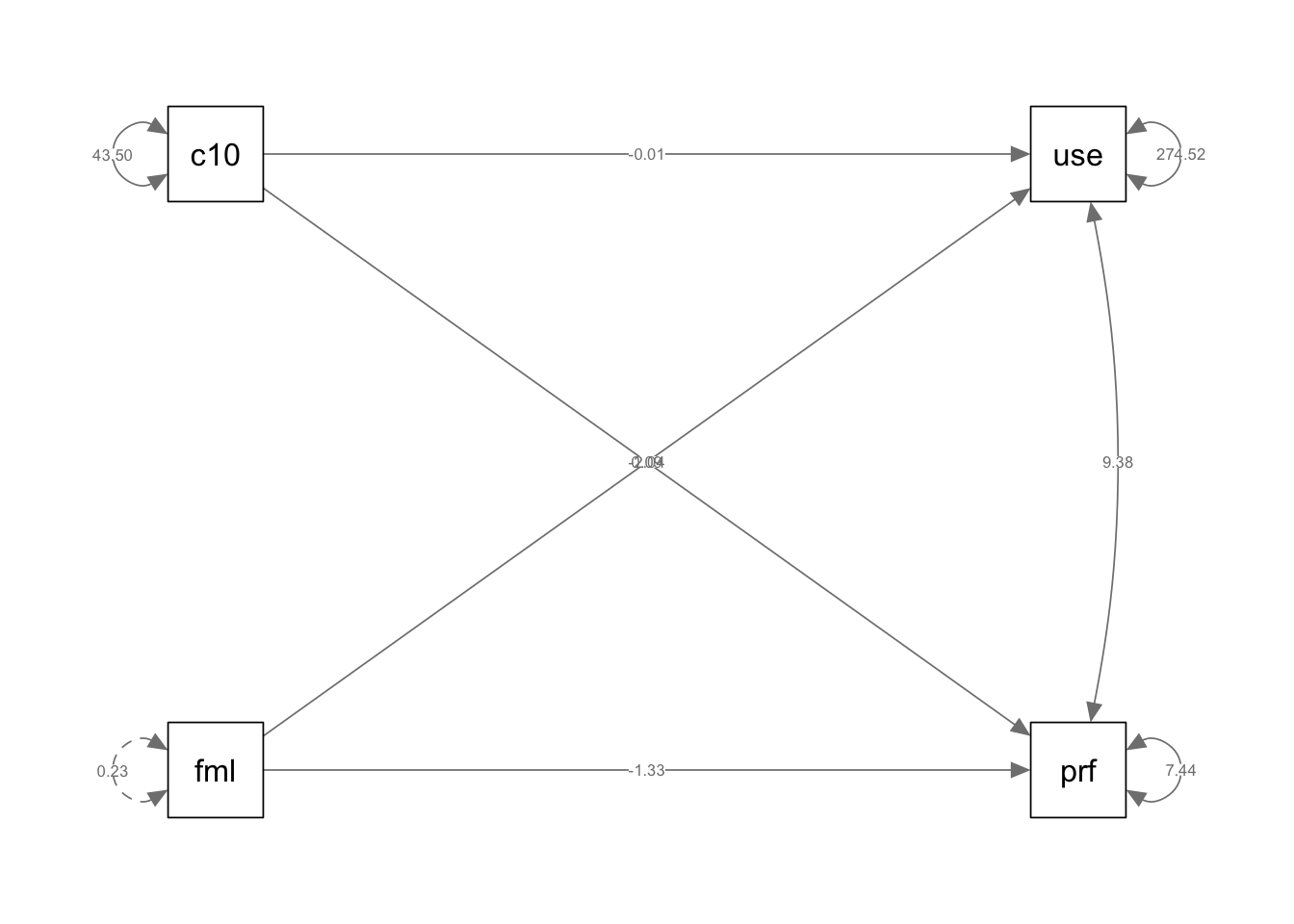
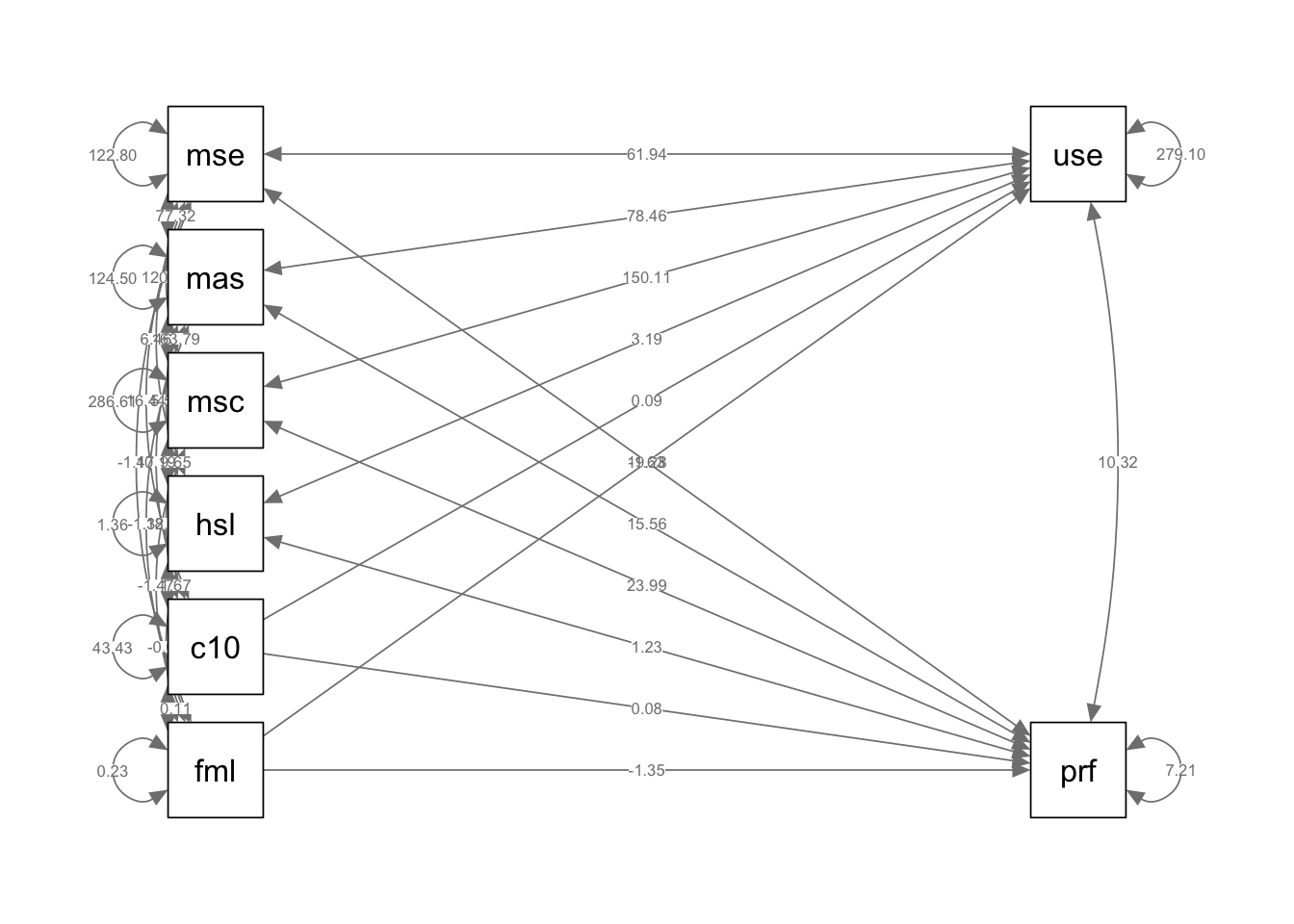
if (!require(mvtnorm)) install.packages("mvtnorm"); library(mvtnorm)Loading required package: mvtnormif (!require(lavaan)) install.packages("lavaan"); library(lavaan)Loading required package: lavaanThis is lavaan 0.6-19
lavaan is FREE software! Please report any bugs.if (!require(semPlot)) install.packages("semPlot"); library(semPlot)Loading required package: semPlot