Bayesian Psychometric Model Fit Methods
Today’s Lecture Objectives
- Show how to use PPMC to evaluate absolute model fit in Bayesian psychometric models
- Show how to use LOO and WAIC for relative model fit in Bayesian psychometric models
Example Data: Conspiracy Theories
Today’s example is from a bootstrap resample of 177 undergraduate students at a large state university in the Midwest. The survey was a measure of 10 questions about their beliefs in various conspiracy theories that were being passed around the internet in the early 2010s. Additionally, gender was included in the survey. All items responses were on a 5- point Likert scale with:
- Strongly Disagree
- Disagree
- Neither Agree or Disagree
- Agree
- Strongly Agree
Please note, the purpose of this survey was to study individual beliefs regarding conspiracies. The questions can provoke some strong emotions given the world we live in currently. All questions were approved by university IRB prior to their use.
Our purpose in using this instrument is to provide a context that we all may find relevant as many of these conspiracy theories are still prevalent today.
Conspiracy Theory Questions 1-5
Questions:
- The U.S. invasion of Iraq was not part of a campaign to fight terrorism, but was driven by oil companies and Jews in the U.S. and Israel.
- Certain U.S. government officials planned the attacks of September 11, 2001 because they wanted the United States to go to war in the Middle East.
- President Barack Obama was not really born in the United States and does not have an authentic Hawaiian birth certificate.
- The current financial crisis was secretly orchestrated by a small group of Wall Street bankers to extend the power of the Federal Reserve and further their control of the world’s economy.
- Vapor trails left by aircraft are actually chemical agents deliberately sprayed in a clandestine program directed by government officials.
Conspiracy Theory Questions 6-10
Questions:
- Billionaire George Soros is behind a hidden plot to destabilize the American government, take control of the media, and put the world under his control.
- The U.S. government is mandating the switch to compact fluorescent light bulbs because such lights make people more obedient and easier to control.
- Government officials are covertly Building a 12-lane "NAFTA superhighway" that runs from Mexico to Canada through America’s heartland.
- Government officials purposely developed and spread drugs like crack-cocaine and diseases like AIDS in order to destroy the African American community.
- God sent Hurricane Katrina to punish America for its sins.
Model Setup Today
Today, we will revert back to the graded response model assumptions to discuss how to estimate the latent variable standard deviation
\[P\left(Y_{ic } = c \mid \theta_p \right) = \left\{ \begin{array}{lr} 1-P\left(Y_{i1} \gt 1 \mid \theta_p \right) & \text{if } c=1 \\ P\left(Y_{i{c-1}} \gt c-1 \mid \theta_p \right) - P\left(Y_{i{c}} \gt c \mid \theta_p \right) & \text{if } 1<c<C_i \\ P\left(Y_{i{C_i -1} } \gt C_i-1 \mid \theta_p \right) & \text{if } c=C_i \\ \end{array} \right.\]
Where:
\[ P\left(Y_{i{c}} > c \mid \theta \right) = \frac{\exp(-\tau_{ic}+\lambda_i\theta_p)}{1+\exp(-\tau_{ic}+\lambda_i\theta_p)}\]
With:
- \(C_i-1\) Ordered thresholds: \(\tau_1 < \tau_2 < \ldots < \tau_{C_i-1}\)
We can convert thresholds to intercepts by multiplying by negative one: \(\mu_c = -\tau_c\)
Model Comparisons: One vs. Two Dimensions
We will fit two models to the data:
- A unidimensional model (see Lecture 4d here)
- A two-dimensionsal model (see Lecture 4e here)
We know from previous lectures that the two-dimensional model had a correlation very close to one
- Such a high correlation made it seem implausible there were two dimensions
Posterior Predictive Model Checking for Absolute Fit in Bayesian Psychometric Models
Psychometric Model PPMC
Psychometric models can use posterior predictive model checking (PPMC) to assess how well they fit the data in an absolute sense
- Each iteration of the chain, each item is simulated using model parameter values from that iteration
- Summary statistics are used to evaluate model fit
- Univariate measures (each item, separately):
- Item mean
- Item \(\chi^2\) (comparing observed data with each simulated data set)
- Not very useful unless:
- Parameters have “obscenely” informative priors
- There are cross-item constraints on some parameters (such as all loadings are equal)
- Bivariate measures (each pair of items)
- For binary data: Tetrachoric correlations
- For polytomous data: Polychoric correlations (but difficult to estimate with small samples), pearson correlations
- For other types of data: Pearson correlations
- Univariate measures (each item, separately):
Problems with PPMC
Problems with PPMC include
- No uniform standard for which statistics to use
- Tetrachoric correlations? Pearson correlations?
- No uniform standard by which data should fit, absolutely
- Jihong Zhang has some work on this topic, though:
- Paper in Structural Equation Modeling
- Dissertation on PPMC with M2 statistics (working on publishing)
- Jihong Zhang has some work on this topic, though:
- No way to determine if a model is overparameterized (too complicated)
- Fit only improves to a limit
Implementing PPMC in Stan (one \(\theta\))
Notes:
- Generated quantities block is where to implement PPMC
- Each type of distribution also has a random number generator
- Here,
ordered_logistic_rnggoes withordered_logistic
- Here,
- Each may have some issue in types of inputs (had to go person-by-person in this block)
- Rather than have Stan calculate statistics, I will do so in R
Implementing PPMC in Stan (two \(\theta\)s)
Notes:
- Very similar to one dimension–just using the syntax from the model block within the
ordered_logistic_rngfunction
PPMC Processing
Stan generated a lot of data–but now we must take it from the format of Stan and process it:
# setting up PPMC
simData = modelOrderedLogit_samples$draws(variables = "simY", format = "draws_matrix")
colnames(simData)
dim(simData)
# set up object for storing each iteration's PPMC data
nPairs = choose(10, 2)
pairNames = NULL
for (col in 1:(nItems-1)){
for (row in (col+1):nItems){
pairNames = c(pairNames, paste0("item", row, "_item", col))
}
}PPMC Calculating in R
PPMCsamples = list()
PPMCsamples$correlation = NULL
PPMCsamples$mean = NULL
# loop over each posterior sample's simulated data
for (sample in 1:nrow(simData)){
# create data frame that has observations (rows) by items (columns)
sampleData = data.frame(matrix(data = NA, nrow = nObs, ncol = nItems))
for (item in 1:nItems){
itemColumns = colnames(simData)[grep(pattern = paste0("simY\\[", item, "\\,"), x = colnames(simData))]
sampleData[,item] = t(simData[sample, itemColumns])
}
# with data frame created, apply functions of the data:
# calculate item means
PPMCsamples$mean = rbind(PPMCsamples$mean, apply(X = sampleData, MARGIN = 2, FUN = mean))
# calculate pearson correlations
temp=cor(sampleData)
PPMCsamples$correlation = rbind(PPMCsamples$correlation, temp[lower.tri(temp)])
}PPMC Results Tabulation in R (Mean)
# next, build distributions for each type of statistic
meanSummary = NULL
# for means
for (item in 1:nItems){
tempDist = ecdf(PPMCsamples$mean[,item])
ppmcMean = mean(PPMCsamples$mean[,item])
observedMean = mean(conspiracyItems[,item])
meanSummary = rbind(
meanSummary,
data.frame(
item = paste0("Item", item),
ppmcMean = ppmcMean,
observedMean = observedMean,
residual = observedMean - ppmcMean,
observedMeanPCT = tempDist(observedMean)
)
)
}PPMC Results Tabulation in R (Correlation)
# for pearson correlations
corrSummary = NULL
# for means
for (column in 1:ncol(PPMCsamples$correlation)){
# get item numbers from items
items = unlist(strsplit(x = colnames(PPMCsamples$correlation)[column], split = "_"))
item1num = as.numeric(substr(x = items[1], start = 5, stop = nchar(items[1])))
item2num = as.numeric(substr(x = items[2], start = 5, stop = nchar(items[2])))
tempDist = ecdf(PPMCsamples$correlation[,column])
ppmcCorr = mean(PPMCsamples$correlation[,column])
observedCorr = cor(conspiracyItems[,c(item1num, item2num)])[1,2]
pct = tempDist(observedCorr)
if (pct > .975 | pct < .025){
inTail = TRUE
} else {
inTail = FALSE
}
corrSummary = rbind(
corrSummary,
data.frame(
item1 = paste0("Item", item1num),
item2 = paste0("Item", item2num),
ppmcCorr = ppmcCorr,
observedCorr = observedCorr,
residual = observedCorr - ppmcCorr,
observedCorrPCT = pct,
inTail = inTail
)
)
}
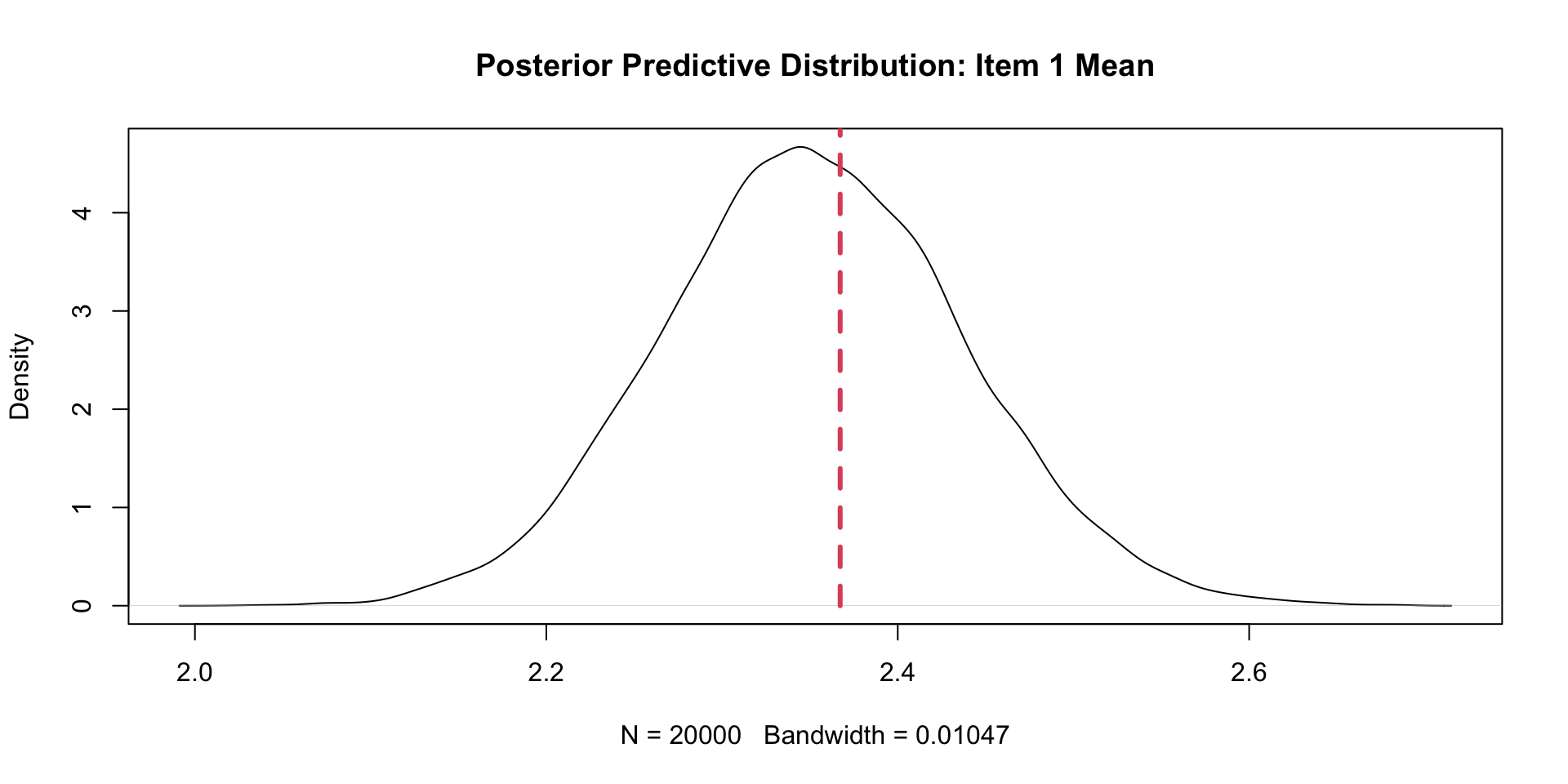
View(corrSummary)PPMC Mean Results: Example Item (One \(\theta\))
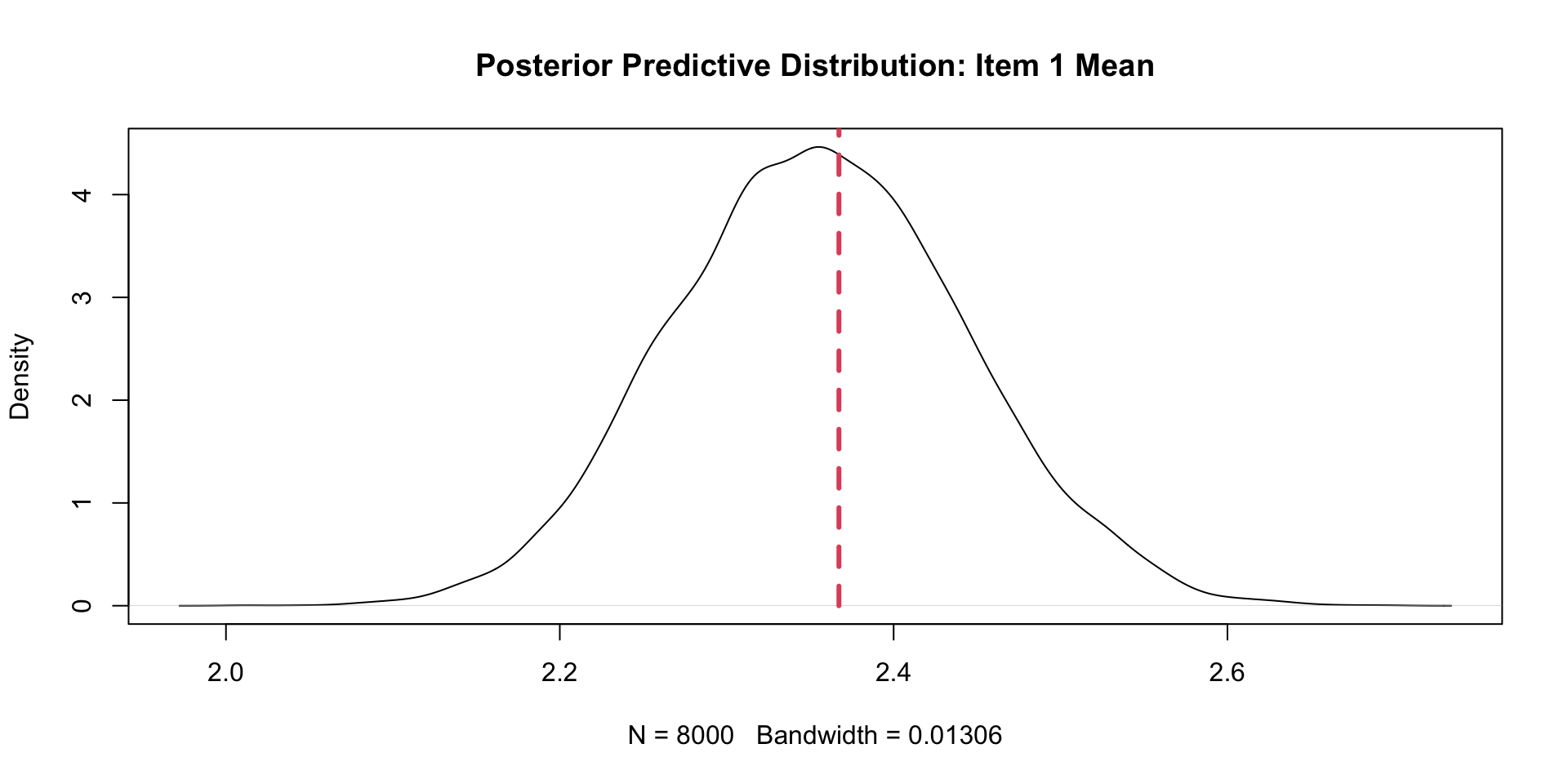
PPMC Mean Results: Example Item (Two \(\theta\)s)
PPMC Mean Results
One \(\theta\)
item ppmcMean observedMean residual observedMeanPCT
1 Item1 2.351961 2.367232 0.015270904 0.58750
2 Item2 1.958019 1.954802 -0.003216949 0.47760
3 Item3 1.862903 1.875706 0.012803672 0.58730
4 Item4 2.007206 2.011299 0.004093220 0.54505
5 Item5 1.978496 1.983051 0.004554802 0.52495
6 Item6 1.880662 1.892655 0.011992938 0.61725
7 Item7 1.748742 1.723164 -0.025578249 0.37475
8 Item8 1.840632 1.841808 0.001175989 0.51105
9 Item9 1.826041 1.807910 -0.018131638 0.42525
10 Item10 1.534652 1.519774 -0.014878249 0.45995Two \(\theta\)s
item ppmcMean observedMean residual observedMeanPCT
1 Item1 2.355617 2.367232 0.0116151130 0.567375
2 Item2 1.966982 1.954802 -0.0121800847 0.425375
3 Item3 1.875570 1.875706 0.0001362994 0.521375
4 Item4 2.017831 2.011299 -0.0065310734 0.487875
5 Item5 1.989239 1.983051 -0.0061878531 0.457625
6 Item6 1.895312 1.892655 -0.0026567797 0.499625
7 Item7 1.758679 1.723164 -0.0355148305 0.325875
8 Item8 1.853364 1.841808 -0.0115557910 0.422500
9 Item9 1.836593 1.807910 -0.0286829096 0.374125
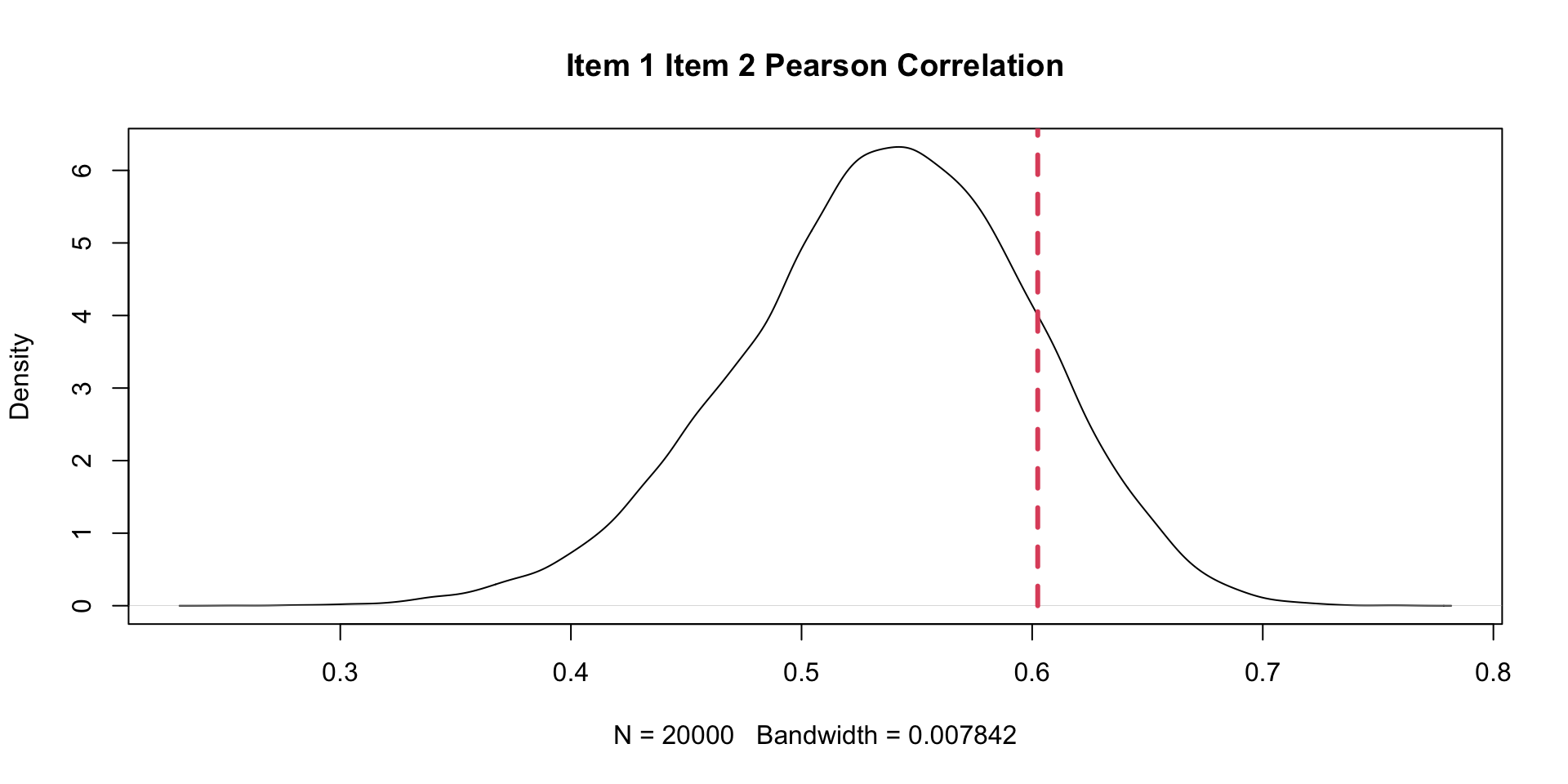
10 Item10 1.552025 1.519774 -0.0322507062 0.376625PPMC Correlation Results: Example Item Pair (One \(\theta\))
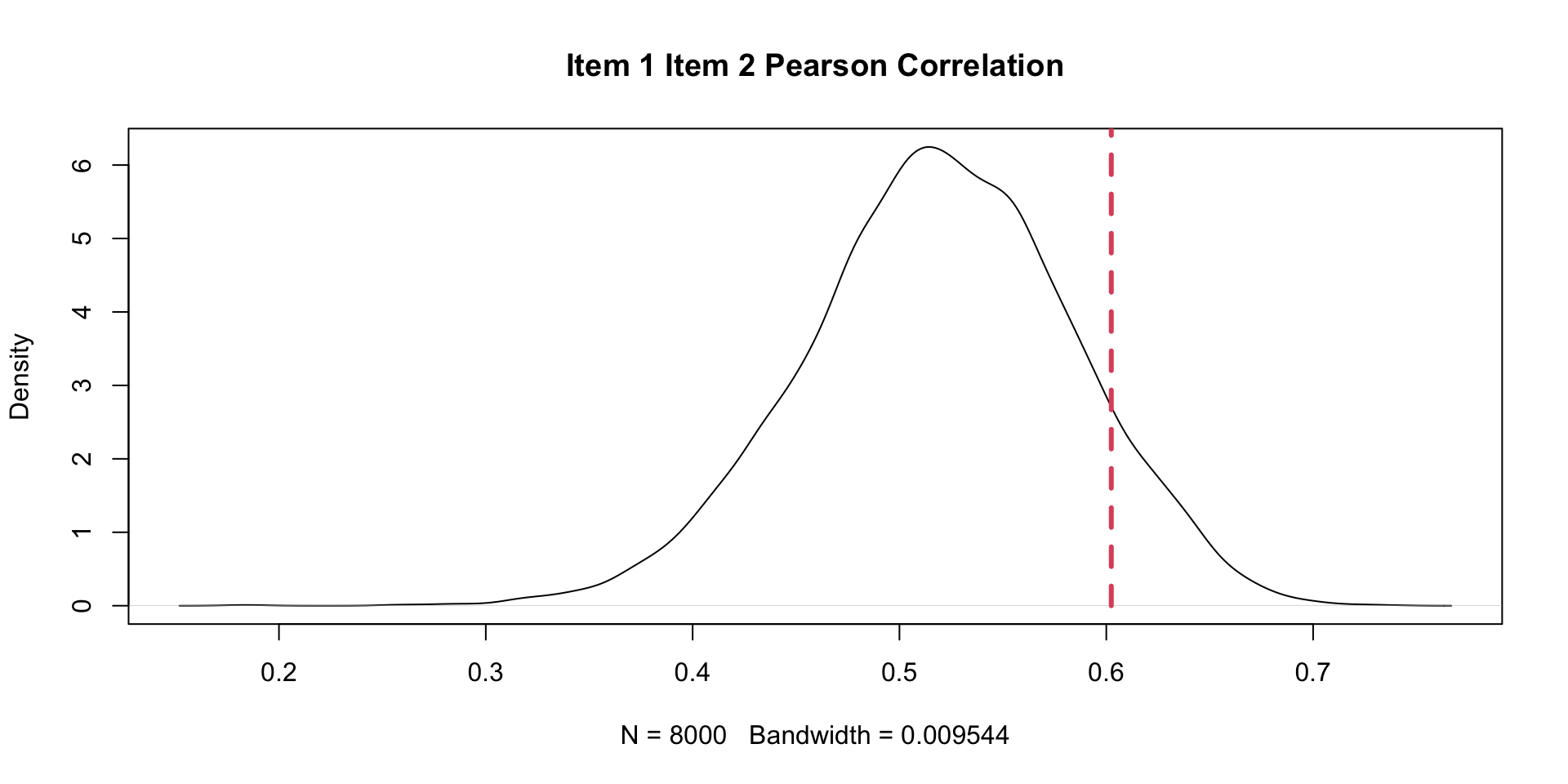
PPMC Correlation Results: Example Item Pair (Two \(\theta\)s)
PPMC Correlation Results
item1 item2 observedCorr ppmcCorr.x residual.x observedCorrPCT.x inTail.x
1 Item10 Item1 0.2568334 0.4138354 -0.157002024 0.01610 TRUE
2 Item10 Item2 0.4121264 0.4922525 -0.080126104 0.13835 FALSE
3 Item10 Item3 0.5670156 0.4528339 0.114181723 0.93580 FALSE
4 Item10 Item4 0.4516124 0.4885576 -0.036945276 0.29200 FALSE
5 Item10 Item5 0.5822203 0.5574134 0.024806856 0.61855 FALSE
6 Item10 Item6 0.5092003 0.5503082 -0.041107914 0.25660 FALSE
7 Item10 Item7 0.4420205 0.4994767 -0.057456184 0.22105 FALSE
8 Item10 Item8 0.5457127 0.5433053 0.002407421 0.48650 FALSE
9 Item10 Item9 0.4244916 0.4940490 -0.069557415 0.18350 FALSE
10 Item2 Item1 0.6024186 0.5359091 0.066509519 0.85330 FALSE
11 Item3 Item1 0.3473072 0.4935820 -0.146274751 0.01650 TRUE
12 Item3 Item2 0.5176819 0.5576809 -0.039998980 0.26080 FALSE
13 Item4 Item1 0.5700901 0.5428928 0.027197305 0.65445 FALSE
14 Item4 Item2 0.5815808 0.6093644 -0.027783572 0.30365 FALSE
15 Item4 Item3 0.4973987 0.5611649 -0.063766138 0.15725 FALSE
16 Item5 Item1 0.5526073 0.6009026 -0.048295276 0.20115 FALSE
17 Item5 Item2 0.6512768 0.6835498 -0.032272935 0.25520 FALSE
18 Item5 Item3 0.5942163 0.6295476 -0.035331390 0.26010 FALSE
19 Item5 Item4 0.6267160 0.6870688 -0.060352830 0.11680 FALSE
20 Item6 Item1 0.5435771 0.5995125 -0.055935394 0.16770 FALSE
21 Item6 Item2 0.6835282 0.6808187 0.002709457 0.49480 FALSE
22 Item6 Item3 0.6601292 0.6278075 0.032321713 0.69385 FALSE
23 Item6 Item4 0.6251455 0.6850634 -0.059917805 0.11470 FALSE
24 Item6 Item5 0.7366890 0.7695588 -0.032869755 0.18350 FALSE
25 Item7 Item1 0.4084389 0.5194172 -0.110978373 0.04575 FALSE
26 Item7 Item2 0.4721628 0.5988614 -0.126698559 0.02615 FALSE
27 Item7 Item3 0.6193269 0.5523662 0.066960776 0.83805 FALSE
28 Item7 Item4 0.4676834 0.5998327 -0.132149297 0.02110 TRUE
29 Item7 Item5 0.6204485 0.6773089 -0.056860368 0.14505 FALSE
30 Item7 Item6 0.6870303 0.6737748 0.013255581 0.57535 FALSE
31 Item8 Item1 0.5425166 0.5986574 -0.056140864 0.16425 FALSE
32 Item8 Item2 0.6317705 0.6778893 -0.046118738 0.17825 FALSE
33 Item8 Item3 0.6182736 0.6274238 -0.009150219 0.41275 FALSE
34 Item8 Item4 0.6373299 0.6837991 -0.046469289 0.17205 FALSE
35 Item8 Item5 0.7641703 0.7677210 -0.003550790 0.43340 FALSE
36 Item8 Item6 0.7557355 0.7699377 -0.014202227 0.33985 FALSE
37 Item8 Item7 0.6611475 0.6720679 -0.010920383 0.39185 FALSE
38 Item9 Item1 0.4420860 0.5122848 -0.070198727 0.14005 FALSE
39 Item9 Item2 0.5998366 0.5915361 0.008300469 0.52950 FALSE
40 Item9 Item3 0.3912734 0.5444218 -0.153148412 0.01820 TRUE
41 Item9 Item4 0.6108052 0.5916837 0.019121483 0.60385 FALSE
42 Item9 Item5 0.7119365 0.6697125 0.042224063 0.76450 FALSE
43 Item9 Item6 0.5849045 0.6646942 -0.079789638 0.07475 FALSE
44 Item9 Item7 0.5653926 0.5911773 -0.025784784 0.32985 FALSE
45 Item9 Item8 0.6068774 0.6612584 -0.054381020 0.14910 FALSE
ppmcCorr.y residual.y observedCorrPCT.y inTail.y
1 0.4061719 -0.149338459 0.019750 TRUE
2 0.4728934 -0.060766997 0.208000 FALSE
3 0.4432168 0.123798859 0.952125 FALSE
4 0.4806637 -0.029051370 0.329250 FALSE
5 0.5325573 0.049662958 0.748750 FALSE
6 0.5329112 -0.023710912 0.345875 FALSE
7 0.4730513 -0.031030770 0.330500 FALSE
8 0.5164191 0.029293644 0.652750 FALSE
9 0.4759515 -0.051459892 0.250625 FALSE
10 0.5187377 0.083680835 0.903750 FALSE
11 0.4844662 -0.137158969 0.023750 TRUE
12 0.5411130 -0.023431145 0.341500 FALSE
13 0.5347878 0.035302267 0.702000 FALSE
14 0.5924472 -0.010866351 0.414125 FALSE
15 0.5549878 -0.057589067 0.182750 FALSE
16 0.5792968 -0.026689512 0.317125 FALSE
17 0.6651563 -0.013879465 0.373625 FALSE
18 0.6074530 -0.013236762 0.389375 FALSE
19 0.6655004 -0.038784360 0.215875 FALSE
20 0.5819181 -0.038341061 0.244625 FALSE
21 0.6519629 0.031565341 0.707625 FALSE
22 0.6102020 0.049927236 0.786375 FALSE
23 0.6680081 -0.042862529 0.195750 FALSE
24 0.7320625 0.004626481 0.522750 FALSE
25 0.4976117 -0.089172822 0.087500 FALSE
26 0.5788783 -0.106715494 0.052125 FALSE
27 0.5283414 0.090985572 0.913000 FALSE
28 0.5765790 -0.108895604 0.046750 FALSE
29 0.6502708 -0.029822324 0.289500 FALSE
30 0.6361468 0.050883503 0.809000 FALSE
31 0.5699207 -0.027404069 0.313750 FALSE
32 0.6513229 -0.019552366 0.342375 FALSE
33 0.5972730 0.021000630 0.615250 FALSE
34 0.6532817 -0.015951841 0.354125 FALSE
35 0.7323234 0.031846868 0.765500 FALSE
36 0.7203063 0.035429192 0.786375 FALSE
37 0.6368085 0.024338967 0.650875 FALSE
38 0.4984311 -0.056345035 0.191250 FALSE
39 0.5802541 0.019582500 0.596125 FALSE
40 0.5294833 -0.138209854 0.028375 FALSE
41 0.5780243 0.032780854 0.686125 FALSE
42 0.6538368 0.058099714 0.844250 FALSE
43 0.6378206 -0.052916032 0.174875 FALSE
44 0.5730831 -0.007690532 0.432375 FALSE
45 0.6377858 -0.030908392 0.274375 FALSECount of Items in Misfitting Pairs
One \(\theta\)
badItems
Item1 Item3 Item10 Item4 Item7 Item9
2 2 1 1 1 1 Two \(\theta\)s
badItems2
Item1 Item10 Item3
2 1 1 Plotting Misfitting Items (One \(\theta\))
Relative Model Fit in Bayesian Psychometric Models
Relative Model Fit in Bayesian Psychometric Models
As with other Bayesian models, we can use WAIC and LOO to compare the model fit of two Bayesian models
- Of note: There is some debate as to whether or not we should marginalize across the latent variables
- We won’t do that here as that would involve a numeric integral
- What is needed: The conditional log likelihood for each observation at each step of the chain
- Here, we have to sum the log likelihood across all items
- There are built-in functions in Stan to do this
Implementing ELPD in Stan (one \(\theta\))
generated quantities{
// for LOO/WAIC:
vector[nObs] personLike = rep_vector(0.0, nObs);
for (item in 1:nItems){
for (obs in 1:nObs){
// calculate conditional data likelihood for LOO/WAIC
personLike[obs] = personLike[obs] + ordered_logistic_lpmf(Y[item, obs] | lambda[item]*theta[obs], thr[item]);
}
}
}Notes:
ordered_logistic_lpmfneeds the observed data to work (first argument)vector[nObs] personLike = rep_vector(0.0, nObs);is needed to set the values to zero at each iteration
Implementing ELPD in Stan (two \(\theta\)s)
generated quantities{
// for LOO/WAIC:
vector[nObs] personLike = rep_vector(0.0, nObs);
for (item in 1:nItems){
for (obs in 1:nObs){
// calculate conditional data likelihood for LOO/WAIC
personLike[obs] = personLike[obs] + ordered_logistic_lpmf(Y[item, obs] | thetaMatrix[obs,]*lambdaMatrix[item,1:nFactors]', thr[item]);
}
}
}Notes:
ordered_logistic_lpmfneeds the observed data to work (first argument)vector[nObs] personLike = rep_vector(0.0, nObs);is needed to set the values to zero at each iteration
Comparing WAIC Values
Computed from 20000 by 177 log-likelihood matrix.
Estimate SE
elpd_waic -1482.8 70.4
p_waic 137.8 5.1
waic 2965.6 140.8
173 (97.7%) p_waic estimates greater than 0.4. We recommend trying loo instead.
Computed from 8000 by 177 log-likelihood matrix.
Estimate SE
elpd_waic -1491.6 68.1
p_waic 145.7 5.0
waic 2983.2 136.1
175 (98.9%) p_waic estimates greater than 0.4. We recommend trying loo instead. Smaller is better, so the unidimensional model wins (but very high SE for both)
Comparing LOO Values
Computed from 20000 by 177 log-likelihood matrix.
Estimate SE
elpd_loo -1541.2 70.9
p_loo 196.2 5.8
looic 3082.4 141.9
------
MCSE of elpd_loo is NA.
MCSE and ESS estimates assume MCMC draws (r_eff in [0.5, 1.3]).
Pareto k diagnostic values:
Count Pct. Min. ESS
(-Inf, 0.7] (good) 6 3.4% 531
(0.7, 1] (bad) 151 85.3% <NA>
(1, Inf) (very bad) 20 11.3% <NA>
See help('pareto-k-diagnostic') for details.
Computed from 8000 by 177 log-likelihood matrix.
Estimate SE
elpd_loo -1543.7 68.3
p_loo 197.8 5.7
looic 3087.3 136.6
------
MCSE of elpd_loo is NA.
MCSE and ESS estimates assume MCMC draws (r_eff in [0.1, 1.3]).
Pareto k diagnostic values:
Count Pct. Min. ESS
(-Inf, 0.7] (good) 14 7.9% 220
(0.7, 1] (bad) 141 79.7% <NA>
(1, Inf) (very bad) 22 12.4% <NA>
See help('pareto-k-diagnostic') for details.LOO seems to (slightly) prefer the two-dimensional model (but by warnings about bad PSIS)
Comparing LOO Values
loo_compare(list(unidimensional = modelOrderedLogit_samples$loo(variables = "personLike"),
twodimensional = modelOrderedLogit2D_samples$loo(variables = "personLike"))) elpd_diff se_diff
unidimensional 0.0 0.0
twodimensional -2.5 5.0 Again, both models are very close in fit
Wrapping Up
Wrapping Up
Model fit is complicated for psychometric models
- Bayesian model fit methods are even more complicated than non-Bayesian methods
- Open area for research!